The yvyD gene of Bacillus subtilis is under dual control of sigmaB and sigmaH
- PMID: 9852014
- PMCID: PMC107773
- DOI: 10.1128/JB.180.24.6674-6680.1998
The yvyD gene of Bacillus subtilis is under dual control of sigmaB and sigmaH
Abstract
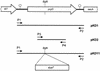
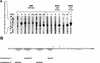
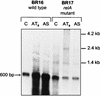
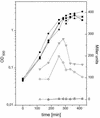
During a search by computer-aided inspection of two-dimensional (2D) protein gels for sigmaB-dependent general stress proteins exhibiting atypical induction profiles, a protein initially called Hst23 was identified as a product of the yvyD gene of Bacillus subtilis. In addition to the typical sigmaB-dependent, stress- and starvation-inducible pattern, yvyD is also induced in response to amino acid depletion. By primer extension of RNA isolated from the wild-type strain and appropriate mutants carrying mutations in the sigB and/or spo0H gene, two promoters were mapped upstream of the yvyD gene. The sigmaB-dependent promoter drives expression of yvyD under stress conditions and after glucose starvation, whereas a sigmaH-dependent promoter is responsible for yvyD transcription following amino acid limitation. Analysis of Northern blots revealed that yvyD is transcribed monocistronically and confirmed the conclusions drawn from the primer extension experiments. The analysis of the protein synthesis pattern in amino acid-starved wild-type and relA mutant cells showed that the YvyD protein is not synthesized in the relA mutant background. It was concluded that the stringent response plays a role in the activation of sigmaH. The yvyD gene product is homologous to a protein which might modify the activity of sigma54 in gram-negative bacteria. The expression of a sigmaL-dependent (sigmaL is the equivalent of sigma54 in B. subtilis) levD-lacZ fusion is upregulated twofold in a yvyD mutant. This indicates that the yvyD gene product, being a member of both the sigmaB and sigmaH regulons, might negatively regulate the activity of the sigmaL regulon. We conclude that (i) systematic, computer-aided analysis of 2D protein gels is appropriate for the identification of genes regulated by multiple transcription factors and that (ii) YvyD might form a junction between the sigmaB and sigmaH regulons on one side and the sigmaL regulon on the other.
Figures



Similar articles
-
The stringent response, sigmaH-dependent gene expression and sporulation in Bacillus subtilis.Mol Gen Genet. 2001 Feb;264(6):913-23. doi: 10.1007/s004380000381. Mol Gen Genet. 2001. PMID: 11254139
-
Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance.Mol Microbiol. 1998 May;28(4):787-802. doi: 10.1046/j.1365-2958.1998.00840.x. Mol Microbiol. 1998. PMID: 9643546
-
Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon.Mol Microbiol. 1998 Sep;29(5):1129-36. doi: 10.1046/j.1365-2958.1998.00977.x. Mol Microbiol. 1998. PMID: 9767581 Review.
-
Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis.Microbiology (Reading). 1999 Apr;145 ( Pt 4):869-880. doi: 10.1099/13500872-145-4-869. Microbiology (Reading). 1999. PMID: 10220166
-
Heat-shock and general stress response in Bacillus subtilis.Mol Microbiol. 1996 Feb;19(3):417-28. doi: 10.1046/j.1365-2958.1996.396932.x. Mol Microbiol. 1996. PMID: 8830234 Review.
Cited by
-
The comER Gene Plays an Important Role in Biofilm Formation and Sporulation in both Bacillus subtilis and Bacillus cereus.Front Microbiol. 2016 Jun 28;7:1025. doi: 10.3389/fmicb.2016.01025. eCollection 2016. Front Microbiol. 2016. PMID: 27446060 Free PMC article.
-
Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus.J Bacteriol. 2008 Jul;190(14):4997-5008. doi: 10.1128/JB.01846-07. Epub 2008 May 16. J Bacteriol. 2008. PMID: 18487332 Free PMC article.
-
The Interaction Network Ontology-supported modeling and mining of complex interactions represented with multiple keywords in biomedical literature.BioData Min. 2016 Dec 19;9:41. doi: 10.1186/s13040-016-0118-0. eCollection 2016. BioData Min. 2016. PMID: 28031747 Free PMC article.
-
Global analysis of the general stress response of Bacillus subtilis.J Bacteriol. 2001 Oct;183(19):5617-31. doi: 10.1128/JB.183.19.5617-5631.2001. J Bacteriol. 2001. PMID: 11544224 Free PMC article.
-
Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.EMBO J. 2017 Jul 14;36(14):2061-2072. doi: 10.15252/embj.201696189. Epub 2017 May 3. EMBO J. 2017. PMID: 28468753 Free PMC article.
References
-
- Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. First steps from a two-dimensional protein index to a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. - PubMed
-
- Belitskii B R, Shakulov R S. Guanosine polyphosphate concentration and stable RNA synthesis in Bacillus subtilis following suppression of protein synthesis. Mol Biol (Moscow) 1980;14:1342–1353. . (In Russian.) - PubMed
-
- Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional gel electrophoresis study. Microbiology. 1997;143:999–1013. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

