Spatial and temporal dynamics of DNA replication sites in mammalian cells
- PMID: 9852140
- PMCID: PMC2132991
- DOI: 10.1083/jcb.143.6.1415
Spatial and temporal dynamics of DNA replication sites in mammalian cells
Abstract
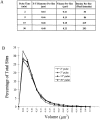
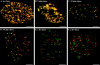
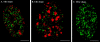
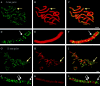
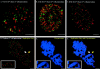
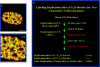
Fluorescence microscopic analysis of newly replicated DNA has revealed discrete granular sites of replication (RS). The average size and number of replication sites from early to mid S-phase suggest that each RS contains numerous replicons clustered together. We are using fluorescence laser scanning confocal microscopy in conjunction with multidimensional image analysis to gain more precise information about RS and their spatial-temporal dynamics. Using a newly improved imaging segmentation program, we report an average of approximately 1,100 RS after a 5-min pulse labeling of 3T3 mouse fibroblast cells in early S-phase. Pulse-chase-pulse double labeling experiments reveal that RS take approximately 45 min to complete replication. Appropriate calculations suggest that each RS contains an average of 1 mbp of DNA or approximately 6 average-sized replicons. Double pulse-double chase experiments demonstrate that the DNA sequences replicated at individual RS are precisely maintained temporally and spatially as the cell progresses through the cell cycle and into subsequent generations. By labeling replicated DNA at the G1/S borders for two consecutive cell generations, we show that the DNA synthesized at early S-phase is replicated at the same time and sites in the next round of replication.
Figures


References
-
- Aten JA, Bakker PJM, Stap J, Boschman GA, Veenhof CHN. DNA double labeling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem J. 1992;24:251–259. - PubMed
-
- Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. - PubMed
-
- Berezney, R. 1984. Organization and functions of the nuclear matrix. In chromosomal nonhistone proteins. L.S. Hnilica, editor. CRC Press Inc., Boca Raton, FL. 4:119–180.
-
- Berezney R. Visualizing DNA replication sites in the cell nucleus. Semin Cell Biol. 1991;2:103–115. - PubMed
-
- Berezney R, Buchholtz LA. Dynamic association of replicating DNA fragments with the nuclear matrix of regenerating liver. Exp Cell Res. 1981;132:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

