A novel nuclear import pathway for the transcription factor TFIIS
- PMID: 9852143
- PMCID: PMC2132971
- DOI: 10.1083/jcb.143.6.1447
A novel nuclear import pathway for the transcription factor TFIIS
Abstract
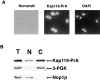
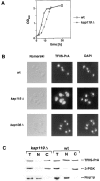
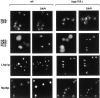
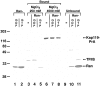
We have identified a novel pathway for protein import into the nucleus. We have shown that the previously identified but uncharacterized yeast protein Nmd5p functions as a karyopherin. It was therefore designated Kap119p (karyopherin with Mr of 119 kD). We localized Kap119p to both the nucleus and the cytoplasm. We identified the transcription elongation factor TFIIS as its major cognate import substrate. The cytoplasmic Kap119p exists as an approximately stoichiometric complex with TFIIS. RanGTP, not RanGDP, dissociated the isolated Kap119p/TFIIS complex and bound to Kap119p. Kap119p also bound directly to a number of peptide repeat containing nucleoporins in overlay assays. In wild-type cells, TFIIS was primarily localized to the nucleus. In a strain where KAP119 has been deleted, TFIIS was mislocalized to the cytoplasm indicating that TFIIS is imported into the nucleus by Kap119p. The transport of various substrates that use other karyopherin-mediated import or export pathways was not affected in a kap119Delta strain. Hence Kap119p is a novel karyopherin that is responsible for the import of the transcription elongation factor TFIIS.
Figures


Similar articles
-
Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting.J Cell Biol. 1999 Jun 28;145(7):1407-17. doi: 10.1083/jcb.145.7.1407. J Cell Biol. 1999. PMID: 10385521 Free PMC article.
-
Role of the nuclear transport factor p10 in nuclear import.Science. 1996 Apr 5;272(5258):120-2. doi: 10.1126/science.272.5258.120. Science. 1996. PMID: 8600522
-
The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins.J Cell Biol. 2001 Feb 19;152(4):729-40. doi: 10.1083/jcb.152.4.729. J Cell Biol. 2001. PMID: 11266464 Free PMC article.
-
Distinct nuclear import and export pathways mediated by members of the karyopherin beta family.J Cell Biochem. 1998 Aug 1;70(2):231-9. J Cell Biochem. 1998. PMID: 9671229 Review.
-
Nuclear import of histones.Biochem Soc Trans. 2020 Dec 18;48(6):2753-2767. doi: 10.1042/BST20200572. Biochem Soc Trans. 2020. PMID: 33300986 Free PMC article. Review.
Cited by
-
Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha.Mol Cell Biol. 2000 Nov;20(22):8468-79. doi: 10.1128/MCB.20.22.8468-8479.2000. Mol Cell Biol. 2000. PMID: 11046143 Free PMC article.
-
Nuclear import of dimerized ribosomal protein Rps3 in complex with its chaperone Yar1.Sci Rep. 2016 Nov 7;6:36714. doi: 10.1038/srep36714. Sci Rep. 2016. PMID: 27819319 Free PMC article.
-
Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus.J Cell Biol. 2005 Dec 19;171(6):967-79. doi: 10.1083/jcb.200504104. J Cell Biol. 2005. PMID: 16365163 Free PMC article.
-
Nuclear import of histone H2A and H2B is mediated by a network of karyopherins.J Cell Biol. 2001 Apr 16;153(2):251-62. doi: 10.1083/jcb.153.2.251. J Cell Biol. 2001. PMID: 11309407 Free PMC article.
-
Transport into and out of the nucleus.Microbiol Mol Biol Rev. 2001 Dec;65(4):570-94, table of contents. doi: 10.1128/MMBR.65.4.570-594.2001. Microbiol Mol Biol Rev. 2001. PMID: 11729264 Free PMC article. Review.
References
-
- Aebi M, Clark MW, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol Gen Genet. 1990;224:72–80. - PubMed
-
- Agarwal K, Baek KH, Jeon CJ, Miyamoto K, Ueno A, Yoon H. Stimulation of transcript elongation requires both the zinc finger and RNA polymerase II binding domains of human TFIIS. Biochemistry. 1991;30:7842–7851. - PubMed
-
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

