Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation
- PMID: 9852144
- PMCID: PMC2132986
- DOI: 10.1083/jcb.143.6.1457
Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation
Abstract
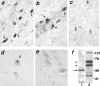
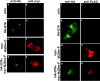
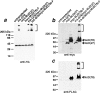
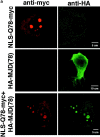
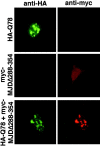
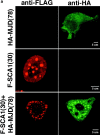
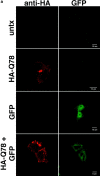
The inherited neurodegenerative diseases caused by an expanded glutamine repeat share the pathologic feature of intranuclear aggregates or inclusions (NI). Here in cell-based studies of the spinocerebellar ataxia type-3 disease protein, ataxin-3, we address two issues central to aggregation: the role of polyglutamine in recruiting proteins into NI and the role of nuclear localization in promoting aggregation. We demonstrate that full-length ataxin-3 is readily recruited from the cytoplasm into NI seeded either by a pathologic ataxin-3 fragment or by a second unrelated glutamine-repeat disease protein, ataxin-1. Experiments with green fluorescence protein/polyglutamine fusion proteins show that a glutamine repeat is sufficient to recruit an otherwise irrelevant protein into NI, and studies of human disease tissue and a Drosophila transgenic model provide evidence that specific glutamine-repeat-containing proteins, including TATA-binding protein and Eyes Absent protein, are recruited into NI in vivo. Finally, we show that nuclear localization promotes aggregation: an ataxin-3 fragment containing a nonpathologic repeat of 27 glutamines forms inclusions only when targeted to the nucleus. Our findings establish the importance of the polyglutamine domain in mediating recruitment and suggest that pathogenesis may be linked in part to the sequestering of glutamine-containing cellular proteins. In addition, we demonstrate that the nuclear environment may be critical for seeding polyglutamine aggregates.
Figures








References
-
- Banif S, Servadio A, Chung M, Kwiatkowski TJ, McCall AE, Duvick LA, Shen Y, Roth EJ, Orr HT, Zoghbi HY. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet. 1994;7:513–520. - PubMed
-
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophilaeye. Cell. 1993;72:379–395. - PubMed
-
- Bonini NM, Leiserson WM, Benzer S. Expression and multiple roles of the eyes absent gene in Drosophila. . Dev Biol. 1998;129:42–57. - PubMed
-
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. - PubMed
-
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

