Gating of skeletal and cardiac muscle sodium channels in mammalian cells
- PMID: 9852324
- PMCID: PMC2269069
- DOI: 10.1111/j.1469-7793.1999.425ae.x
Gating of skeletal and cardiac muscle sodium channels in mammalian cells
Abstract
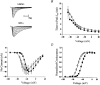
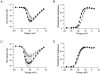
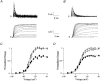
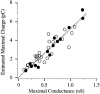
1. Sodium channel ionic current (INa) and gating current (Ig) were compared for rat skeletal (rSkM1) and human heart Na+ channels (hH1a) heterologously expressed in cultured mammalian cells at approximately 13 C before and after modification by site-3 toxins (Anthopleurin A and Anthopleurin B). 2. For hH1a Na+ channels there was a concordance between the half-points (V ) of the peak conductance-voltage (G-V) relationship and the gating charge-voltage (Q-V) relationship with no significant difference in half-points. In contrast, the half-point of the Q-V relationship for rSkM1 Na+ channels was shifted to more negative potentials compared with its G-V relationship with a significant difference in the half-points of -8 mV. 3. Site-3 toxins slowed the decay of INa in response to step depolarizations for both rSkM1 and hH1a Na+ channels. The half-point of the G-V relationship in rSkM1 Na+ channels was shifted by -8.0 mV while toxin modification of hH1a Na+ channels produced a smaller hyperpolarizing shift of the V by -3.7 mV. 4. Site-3 toxins reduced maximal gating charge (Qmax ) by 33% in rSkM1 and by 31% in hH1a, but produced only minor changes in the half-points and slope factors of their Q-V relationships. In contrast to measurements in control solutions, after modification by site-3 toxin the half-points of the G-V and the Q-V relationships for rSkM1 Na+ channels demonstrated a concordance similar to that for hH1a. 5. Qmax vs. Gmax for rSkM1 and hH1a Na+ channels exhibited linear relationships with almost identical slopes, as would be expected if the number of electronic charges (e-) per channel was comparable. 6. We conclude that the faster kinetics in rSkM1 channels compared with hH1a channels may arise from inherently faster rate transitions in skeletal muscle Na+ channels, and not from major differences in the voltage dependence of the channel transitions.
Figures
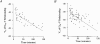
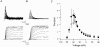
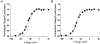

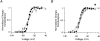
References
-
- Aldrich RW, Corey DP, Stevens CF. A reinterpretation of mammalian sodium channel gating based on single channel recording. Nature. 1983;306:436–441. - PubMed
-
- Benzinger GR, Drum CL, Chen L-Q, Kallen RG, Hanck DA. Differences in the binding sites of two site-3 sodium channel toxins. Pflügers Archiv. 1997;434:742–749. - PubMed
-
- Chahine M, Deschene I, Chen LQ, Kallen RG. Electrophysiological characteristics of cloned skeletal and cardiac muscle sodium channels. American Journal of Physiology. 1996;271:H498–506. - PubMed
-
- Conti F, Stühmer W. Quantal charge redistribution accompanying the structural transition of sodium channels. European Biophysical Journal. 1989;17:53–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

