Helper-free foamy virus vectors
- PMID: 9853518
- PMCID: PMC3010407
- DOI: 10.1089/hum.1998.9.17-2517
Helper-free foamy virus vectors
Abstract
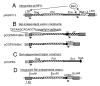
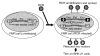
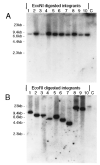
Retroviral vectors based on human foamy virus (HFV) have been developed and show promise as gene therapy vehicles. Here we describe a method for the production of HFV vector stocks free of detectable helper virus. The helper and vector plasmid constructs used both lack the HFV bel genes, so recombination between these constructs cannot create a wild-type virus. A fusion promoter that combines portions of the cytomegalovirus (CMV) immediate-early and HFV long terminal repeat (LTR) promoters was used to drive expression of both the helper and vector constructs. The CMV-LTR fusion promoter allows for HFV vector production in the absence of the Bel-1 trans-activator protein, which would otherwise be necessary for efficient transcription from the HFV LTR. Vector stocks containing either neomycin phosphotransferase or alkaline phosphatase reporter genes were produced by transient transfection at titers greater than 10(5) transducing units/ml. G418-resistant BHK-21 cells obtained by transduction with neo vectors contained randomly integrated HFV vector proviruses without detectable deletions or rearrangements. The vector stocks generated were free of replication-competent retrovirus (RCR), as determined by assays for LTR trans-activation and a marker rescue assay developed here for the detection of Bel-independent RCR.
Figures



References
-
- ACHONG BG, MANSELL PW, EPSTEIN MA, CLIFFORD P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 1971;46:299–307. - PubMed
-
- BIENIASZ PD, ERLWEIN O, AGUZZI A, RETHWILM A, MCCLURE MO. Gene transfer using replication-defective human foamy virus vectors. Virology. 1997;235:65–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

