A systematic molecular genetic approach to study mammalian germline development
- PMID: 9853837
- PMCID: PMC3049814
A systematic molecular genetic approach to study mammalian germline development
Abstract
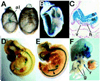
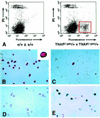
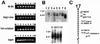
It is difficult to study gene expression in mammalian embryonic germ cells as PGCs constitute only a minor proportion of the mouse embryo. We have overcome this problem by using a novel combination of established molecular and transgenic approaches. A line of mice has been generated in which the cells of the germ lineage express the beta-galactosidase reporter gene during embryogenesis. Using this line, germ cells have been purified to near homogeneity from embryos at discrete stages during germline development by use of a stain for beta-gal activity and a fluorescence activated cell sorter. Subsequently, cDNA libraries have been constructed from each germ cell population using a modified lone-linker PCR strategy. These combined cDNA libraries represent genes expressed in PGCs during mammalian germline development. To facilitate a molecular genetic approach to studying mammalian germline development, these cDNA libraries will be pooled to form an arrayed, addressed reference embryonic germ cell cDNA library. In parallel with large-scale cDNA sequencing efforts; genes that are differentially expressed in germ cells will be identified by screening the reference library with probes generated by subtractive hybridization. Complementary DNAs identified using this approach will be analyzed by sequencing, database comparison, genomic mapping and in situ hybridization to ascertain the potential functional importance of each gene to germline development. In addition to providing a wealth of novel information regarding patterns of gene expression during mammalian germline development, these results will form the basis for future experiments to determine the function of these genes in this process.
Figures


References
-
- Aaronson JS, Eckman B, Blevins RA, Borkowski JA, Myerson J, Imran S, Elliston KO. Toward the development of a gene index to the human genome: an assessment of the nature of high-throughput EST sequence data. Genome Res. 1996;6:829–845. - PubMed
-
- Abe K. Rapid isolation of desired sequences from lone linker PCR amplified cDNA mixtures: Application to identification and recovery of expressed sequences in cloned genomic DNA. Mammal. Genome. 1992;2:252–259. - PubMed
-
- Abe K, Hashiyama M, MacGregor G, Yamamura K-I, Abe K. Purification of primordial germ cells from TNAPβ-geo mouse embryos using FACS-gal. Dev. Biol. 1996;180:468–472. - PubMed
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Alvarez-Buylla A, Merchant-Larios H. Mouse primordial germ cells use fibronectin as a substrate for migration. Exp. Cell Res. 1986;165:362–368. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
