The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells
- PMID: 9858508
- PMCID: PMC2212439
- DOI: 10.1084/jem.188.12.2215
The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells
Abstract
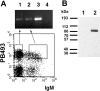
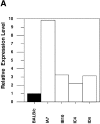
In mature B lymphocytes, the zinc finger transcription factor early growth response 1 (Egr-1) is one of the many immediate-early genes induced upon B cell antigen receptor engagement. However, its role during earlier stages of lymphopoiesis has remained unclear. By examining bone marrow B cell subsets, we found Egr-1 transcripts in pro/pre-B and immature B lymphocytes, and Egr-1 protein in pro/pre-B-I cells cultivated on stroma cells in the presence of interleukin (IL)-7. In recombinase-activating gene (RAG)-2-deficient mice overexpressing an Egr-1 transgene in the B lymphocyte lineage, pro/pre-B-I cells could differentiate past a developmental block at the B220(low) BP-1(-) stage to the stage of B220(low) BP-1(+) pre-B-I cells, but not further to the B220(low) BP-1(+) CD25(+) stage of pre-B-II cells. Therefore, during early B lymphopoiesis progression from the B220(low) BP-1(-) IL-2R- pro/pre-B-I stage to the B220(low) BP-1(+) IL-2R+ pre-B-II stage seems to occur in at least two distinct steps, and the first step to the stage of B220(low) BP-1(+) pre-B-I cells can be promoted by the overexpression of Egr-1 alone. Wild-type mice expressing an Egr-1 transgene had increased proportions of mature immunoglobulin (Ig)M+ B220(high) and decreased proportions of immature IgM+ B220(low) bone marrow B cells. Since transgenic and control precursor B cells show comparable proliferation patterns, overexpression of Egr-1 seems also to promote entry into the mature B cell stage. Analysis of changes in the expression pattern of potential Egr-1 target genes revealed that Egr-1 enhances the expression of the aminopeptidase BP-1/6C3 in pre-B and immature B cells and upregulates expression of the orphan nuclear receptor nur77 in IgM+ B cells.
Figures


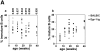
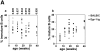


References
-
- Sukhatme VP, Kartha S, Toback FG, Taub R, Hoover RG, Tsai-Morris CH. A novel early growth response gene rapidly induced by fibroblast, epithelial cell and lymphocyte mitogens. Oncogene Res. 1987;1:343–355. - PubMed
-
- Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

