Genetic regulation of commitment to interleukin 4 production by a CD4(+) T cell-intrinsic mechanism
- PMID: 9858515
- PMCID: PMC2212433
- DOI: 10.1084/jem.188.12.2289
Genetic regulation of commitment to interleukin 4 production by a CD4(+) T cell-intrinsic mechanism
Abstract
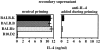
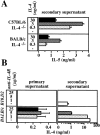
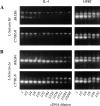
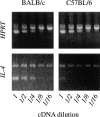
The dysregulated expression of interleukin 4 (IL-4) can have deleterious effects on the outcome of infectious and allergic diseases. Despite this, the mechanisms by which naive T cells commit to IL-4 expression during differentiation into mature effector cells remain incompletely defined. As compared to cells from most strains of mice, activated CD4(+) T cells from BALB mice show a bias towards IL-4 production and T helper 2 commitment in vitro and in vivo. Here, we show that this bias arises not from an increase in the amount of IL-4 produced per cell, but rather from an increase in the proportion of CD4(+) T cells that commit to IL-4 expression. This strain-specific difference in commitment was independent of signals mediated via the IL-4 receptor and hence occurred upstream of potential autoregulatory effects of IL-4. Segregation analysis of the phenotype in an experimental backcross cohort implicated a polymorphic locus on chromosome 16. Consistent with a role in differentiation, expression of the phenotype was CD4(+) T cell intrinsic and was evident as early as 16 h after the activation of naive T cells. Probabilistic gene activation is proposed as a T cell-intrinsic mechanism capable of modulating the proportion of naive T cells that commit to IL-4 production.
Figures





References
-
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

