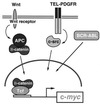
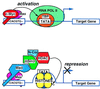
c-Myc target genes involved in cell growth, apoptosis, and metabolism
- PMID: 9858526
- PMCID: PMC83860
- DOI: 10.1128/MCB.19.1.1
c-Myc target genes involved in cell growth, apoptosis, and metabolism
Figures


References
-
- Adams J M, Cory S. Transgenic models of tumor development. Science. 1991;254:1161–1167. - PubMed
-
- Afar D E, Goga A, McLaughlin J, Witte O N, Sawyers C L. Differential complementation of Bcr-Abl point mutants with c-Myc. Science. 1994;264:424–426. - PubMed
-
- Albarosa R, DiDonato S, Finocchiaro G. Redefinition of the coding sequence of the MXI1 gene and identification of a polymorphic repeat in the 3′ non-coding region that allows the detection of loss of heterozygosity of chromosome 10q25 in glioblastomas. Hum Genet. 1995;95:709–711. - PubMed
-
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:D250–D268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
