RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity
- PMID: 9858531
- PMCID: PMC83865
- DOI: 10.1128/MCB.19.1.57
RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity
Abstract
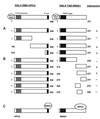
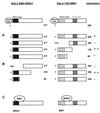
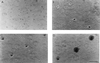
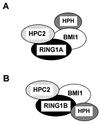
Polycomb-group (PcG) proteins form large multimeric protein complexes that are involved in maintaining the transcriptionally repressive state of genes. Previously, we reported that RING1 interacts with vertebrate Polycomb (Pc) homologs and is associated with or is part of a human PcG complex. However, very little is known about the role of RING1 as a component of the PcG complex. Here we undertake a detailed characterization of RING1 protein-protein interactions. By using directed two-hybrid and in vitro protein-protein analyses, we demonstrate that RING1, besides interacting with the human Pc homolog HPC2, can also interact with itself and with the vertebrate PcG protein BMI1. Distinct domains in the RING1 protein are involved in the self-association and in the interaction with BMI1. Further, we find that the BMI1 protein can also interact with itself. To better understand the role of RING1 in regulating gene expression, we overexpressed the protein in mammalian cells and analyzed differences in gene expression levels. This analysis shows that overexpression of RING1 strongly represses En-2, a mammalian homolog of the well-characterized Drosophila PcG target gene engrailed. Furthermore, RING1 overexpression results in enhanced expression of the proto-oncogenes c-jun and c-fos. The changes in expression levels of these proto-oncogenes are accompanied by cellular transformation, as judged by anchorage-independent growth and the induction of tumors in athymic mice. Our data demonstrate that RING1 interacts with multiple human PcG proteins, indicating an important role for RING1 in the PcG complex. Further, deregulation of RING1 expression leads to oncogenic transformation by deregulation of the expression levels of certain oncogenes.
Figures






References
-
- Alkema M J, Bronk M, Verhoeven E, Otte A P, van’t Veer L J, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. - PubMed
-
- Alkema M J, Jacobs H, Van Lohuizen M, Berns A. Perturbation of B and T cell development and predisposition to lymphomagenesis in EμBmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 1997;15:899–910. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
