Spi-1/PU.1 is a positive regulator of the Fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines
- PMID: 9858537
- PMCID: PMC83871
- DOI: 10.1128/MCB.19.1.121
Spi-1/PU.1 is a positive regulator of the Fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines
Abstract
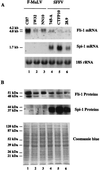
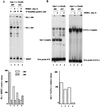
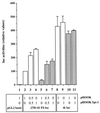
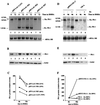
Spi-1/PU.1 and Fli-1 are two members of the ETS family of transcription factors whose expression is deregulated by proviral insertion in most erythroleukemic cell lines induced by the spleen focus-forming virus (SFFV) and Friend murine leukemia virus (F-MuLV) components of the Friend viral complex, respectively. In this study, we present evidence that transcription of the Fli-1 gene is positively regulated by Spi-1/PU.1 in SFFV-transformed cell lines: (i) all SFFV-transformed cell lines expressing Spi-1/PU.1 are characterized by a specific pattern of Fli-1 gene transcripts initiated in the -200 region instead of position -400 as reported for F-MuLV-transformed cell lines; (ii) these Fli-1 transcripts initiated in the -200 region are downregulated in parallel with that of Spi-1/PU.1 during hexamethylenebisacetamide (HMBA) induced differentiation; and (iii) Fli-1 transcription is upregulated in SFFV cells lines following stable transfection of a Spi-1/PU.1 expression vector. Furthermore, we found by transient transfection assays that the -270/-41 region of the Fli-1 gene displays promoter activity which is transactivated by Spi-1/PU.1. This promoter is strictly dependent on the integrity of two highly conserved ETS DNA binding sites that bind the Spi-1/PU.1 protein in vitro. Finally, we show that transfection of constitutive or inducible Fli-1 expression vectors in SFFV-transformed cells inhibits their erythroid differentiation induced by HMBA. Overall, these data indicate that Fli-1 is a target gene of the Spi-1/PU.1 transcription factor in SFFV-transformed cell lines. We further suggest that deregulated synthesis of Fli-1 may trigger a common mechanism contributing to erythroleukemia induced by either SFFV or F-MuLV.
Figures





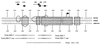


References
-
- Barbeau B, Bergeron D, Beaulieu M, Nadjem Z, Rassart E. Characterization of the human and mouse Fli-1 promoter regions. Biochem Biophys Acta. 1996;1307:220–232. - PubMed
-
- Ben-David Y, Giddens E R, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–919. - PubMed
-
- Ben-David Y, Bernstein A. Friend virus induced erythroleukemia and the multi stage of cancer. Cell. 1991;66:831–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
