The B29 (immunoglobulin beta-chain) gene is a genetic target for early B-cell factor
- PMID: 9858563
- PMCID: PMC83897
- DOI: 10.1128/MCB.19.1.392
The B29 (immunoglobulin beta-chain) gene is a genetic target for early B-cell factor
Abstract
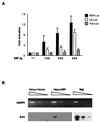
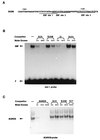
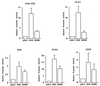
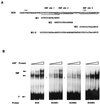
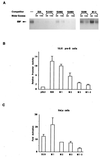
Early B-cell factor (EBF) is a transcription factor suggested as essential for early B-lymphocyte development by findings in mice where the coding gene has been inactivated by homologous disruption. This makes the identification of genetic targets for this transcription factor pertinent for the understanding of early B-cell development. The lack of B29 transcripts, coding for the beta subunit of the B-cell receptor complex, in pro-B cells from EBF-deficient mice suggested that B29 might be a genetic target for EBF. We here present data suggesting that EBF interacts with three independent sites within the mouse B29 promoter. Furthermore, ectopic expression of EBF in HeLa cells activated a B29 promoter-controlled reporter construct 13-fold and induced a low level of expression from the endogenous B29 gene. Finally, mutations in the EBF binding sites diminished B29 promoter activity in pre-B cells while the same mutations did not have as striking an effect on the promoter function in B-cell lines of later differentiation stages. These data suggest that the B29 gene is a genetic target for EBF in early B-cell development.
Figures


References
-
- Åkerblad P, Sigvardsson M, Leanderson T. Early B cell factor (EBF) down regulates immunoglobulin heavy chain intron enhancer function in a plasmacytoma cell line. Scand J Immunol. 1996;44:145–149. - PubMed
-
- Åkerblad, P. Unpublished data.
-
- Bain G, Robanus Maandag E, Izon D, Amsen D, Kruisbeek A, Weintraub B, Krop I, Schlissel M, Feeney A, van Roon M, van der Valk M, te Riele H, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. - PubMed
-
- Borst J, Jacobs H, Brouns G. Composition and function of T cell receptor and B cell receptor complexes on precursor lymphocytes. Curr Opin Immunol. 1996;8:181–190. - PubMed
-
- Clevers H, Grosschedl R. Transcriptional control of lymphoid development: lessons from gene targeting. Immunol Today. 1996;17:336–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
