Testing for DNA tracking by MOT1, a SNF2/SWI2 protein family member
- PMID: 9858565
- PMCID: PMC83899
- DOI: 10.1128/MCB.19.1.412
Testing for DNA tracking by MOT1, a SNF2/SWI2 protein family member
Abstract
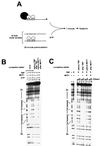
Proteins in the SNF2/SWI2 family use ATP hydrolysis to catalyze rearrangements in diverse protein-DNA complexes. How ATP hydrolysis is coupled to these rearrangements is unknown, however. One attractive model is that these ATPases are ATP-dependent DNA-tracking enzymes. This idea was tested for the SNF2/SWI2 protein family member MOT1. MOT1 is an essential Saccharomyces cerevisiae transcription factor that uses ATP to dissociate TATA binding protein (TBP) from DNA. By using a series of DNA templates with one or two TATA boxes in combination with binding sites for heterologous DNA binding "roadblock" proteins, the ability of MOT1 to track along DNA was assayed. The results demonstrate that, following ATP-dependent TBP-DNA dissociation, MOT1 dissociates rapidly from the DNA by a mechanism that does not require a DNA end. Template commitment footprinting experiments support the conclusion that ATP-dependent DNA tracking by MOT1 does not occur. These results support a model in which MOT1 drives TBP-DNA dissociation by a mechanism that involves a transient, ATP-dependent interaction with TBP-DNA which does not involve ATP-dependent DNA tracking.
Figures






References
-
- Auble, D. T. Unpublished observations.
-
- Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. - PubMed
-
- Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
