Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation
- PMID: 9858581
- PMCID: PMC83915
- DOI: 10.1128/MCB.19.1.577
Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation
Abstract
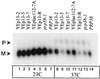
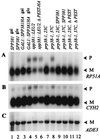
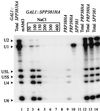
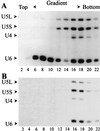
U4 snRNA release from the spliceosome occurs through an essential but ill-defined Prp38p-dependent step. Here we report the results of a dosage suppressor screen to identify genes that contribute to PRP38 function. Elevated expression of a previously uncharacterized gene, SPP381, efficiently suppresses the growth and splicing defects of a temperature-sensitive (Ts) mutant prp38-1. This suppression is specific in that enhanced SPP381 expression does not alter the abundance of intronless RNA transcripts or suppress the Ts phenotypes of other prp mutants. Since SPP381 does not suppress a prp38::LEU2 null allele, it is clear that Spp381p assists Prp38p in splicing but does not substitute for it. Yeast SPP381 disruptants are severely growth impaired and accumulate unspliced pre-mRNA. Immune precipitation studies show that, like Prp38p, Spp381p is present in the U4/U6.U5 tri-snRNP particle. Two-hybrid analyses support the view that the carboxyl half of Spp381p directly interacts with the Prp38p protein. A putative PEST proteolysis domain within Spp381p is dispensable for the Spp381p-Prp38p interaction and for prp38-1 suppression but contributes to Spp381p function in splicing. Curiously, in vitro, Spp381p may not be needed for the chemistry of pre-mRNA splicing. Based on the in vivo and in vitro results presented here, we propose that two small acidic proteins without obvious RNA binding domains, Spp381p and Prp38p, act in concert to promote U4/U5.U6 tri-snRNP function in the spliceosome cycle.
Figures




References
-
- Barnes J A, Gomes A V. PEST sequences in calmodulin-binding proteins. Mol Cell Biochem. 1995;149/150:17–27. - PubMed
-
- Beickman, K., and B. C. Rymond. Unpublished data.
-
- Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
