Cell cycle-dependent regulation of human DNA polymerase alpha-primase activity by phosphorylation
- PMID: 9858588
- PMCID: PMC83922
- DOI: 10.1128/MCB.19.1.646
Cell cycle-dependent regulation of human DNA polymerase alpha-primase activity by phosphorylation
Abstract
DNA polymerase alpha-primase is known to be phosphorylated in human and yeast cells in a cell cycle-dependent manner on the p180 and p68 subunits. Here we show that phosphorylation of purified human DNA polymerase alpha-primase by purified cyclin A/cdk2 in vitro reduced its ability to initiate simian virus 40 (SV40) DNA replication in vitro, while phosphorylation by cyclin E/cdk2 stimulated its initiation activity. Tryptic phosphopeptide mapping revealed a family of p68 peptides that was modified well by cyclin A/cdk2 and poorly by cyclin E/cdk2. The p180 phosphopeptides were identical with both kinases. By mass spectrometry, the p68 peptide family was identified as residues 141 to 160. Cyclin A/cdk2- and cyclin A/cdc2-modified p68 also displayed a phosphorylation-dependent shift to slower electrophoretic mobility. Mutation of the four putative phosphorylation sites within p68 peptide residues 141 to 160 prevented its phosphorylation by cyclin A/cdk2 and the inhibition of replication activity. Phosphopeptide maps of the p68 subunit of DNA polymerase alpha-primase from human cells, synchronized and labeled in G1/S and in G2, revealed a cyclin E/cdk2-like pattern in G1/S and a cyclin A/cdk2-like pattern in G2. The slower-electrophoretic-mobility form of p68 was absent in human cells in G1/S and appeared as the cells entered G2/M. Consistent with this, the ability of DNA polymerase alpha-primase isolated from synchronized human cells to initiate SV40 replication was maximal in G1/S, decreased as the cells completed S phase, and reached a minimum in G2/M. These results suggest that the replication activity of DNA polymerase alpha-primase in human cells is regulated by phosphorylation in a cell cycle-dependent manner.
Figures

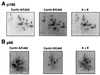
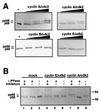

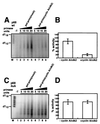

References
-
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. - PubMed
-
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Braun K A, Lao Y, He Z, Ingles C J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RP-A): RP-A modulates the activity of DNA polymerase α by multiple mechanisms. Biochemistry. 1997;36:8443–8454. - PubMed
-
- Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
