CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity
- PMID: 9858599
- PMCID: PMC83933
- DOI: 10.1128/MCB.19.1.764
CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity
Abstract
Genes encoding the Phe-Gly (FG) repeat-containing nucleoporins NUP98 and CAN/NUP214 are at the breakpoints of several chromosomal translocations associated with human acute myeloid leukemia (AML), but their role in oncogenesis is unclear. Here we demonstrate that the NUP98-HOXA9 fusion gene encodes two nuclear oncoproteins with either 19 or 37 NUP98 FG repeats fused to the DNA binding and PBX heterodimerization domains of the transcription factor HOXA9. Both NUP98-HOXA9 chimeras transformed NIH 3T3 fibroblasts, and this transformation required the HOXA9 domains for DNA binding and PBX interaction. Surprisingly, the FG repeats acted as very potent transactivators of gene transcription. This NUP98-derived activity is essential for transformation and can be replaced by the bona fide transactivation domain of VP16. Interestingly, FG repeat-containing segments derived from the nucleoporins NUP153 and CAN/NUP214 functioned similarly to those from NUP98. We further demonstrate that transactivation by FG repeat-rich segments of NUP98 correlates with their ability to interact functionally and physically with the transcriptional coactivators CREB binding protein (CBP) and p300. This finding shows, for the first time, that a translocation-generated fusion protein appears to recruit CBP/p300 as an important step of its oncogenic mechanism. Together, our results suggest that NUP98-HOXA9 chimeras are aberrant transcription factors that deregulate HOX-responsive genes through the transcriptional activation properties of nucleoporin-specific FG repeats that recruit CBP/p300. Indeed, FG repeat-mediated transactivation may be a shared pathogenic function of nucleoporins implicated human AML.
Figures







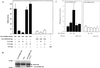

References
-
- Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. - PubMed
-
- Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, Kaneko Y, Ohki M. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–3944. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
