RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast
- PMID: 9858617
- PMCID: PMC83951
- DOI: 10.1128/MCB.19.1.934
RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast
Abstract
The nucleotide excision repair (NER) pathway is able to remove a wide variety of structurally unrelated lesions from DNA. NER operates throughout the genome, but the efficiencies of lesion removal are not the same for different genomic regions. Even within a single gene or DNA strand repair rates vary, and this intragenic heterogeneity is of considerable interest with respect to the mutagenic potential of carcinogens. In this study, we have analyzed the removal of the two major types of genotoxic DNA adducts induced by UV light, i.e., the pyrimidine (6-4)-pyrimidone photoproduct (6-4PP) and the cyclobutane pyrimidine dimer (CPD), from the Saccharomyces cerevisiae URA3 gene at nucleotide resolution. In contrast to the fast and uniform removal of CPDs from the transcribed strand, removal of lesions from the nontranscribed strand is generally less efficient and is modulated by the chromatin environment of the damage. Removal of 6-4PPs from nontranscribed sequences is also profoundly influenced by positioned nucleosomes, but this type of lesion is repaired at a much higher rate. Still, the transcribed strand is repaired preferentially, indicating that, as in the removal of CPDs, transcription-coupled repair predominates in the removal of 6-4PPs from transcribed DNA. The hypothesis that transcription machinery operates as the rate-determining damage recognition entity in transcription-coupled repair is supported by the observation that this pathway removes both types of UV photoproducts at equal rates without being profoundly influenced by the sequence or chromatin context.
Figures
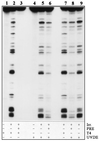


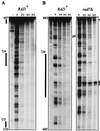

References
-
- Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Produst V N, Protić M, Hübscher U, Egly J-M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology; 1995.
-
- Gao S, Drouin R, Holmquist G P. DNA repair rates mapped along the human PGK1 gene at nucleotide resolution. Science. 1994;263:1438–1440. - PubMed
-
- Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. - PubMed
-
- Guzder S N, Sung P, Prakash L, Prakash S. Yeast Rad7-Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J Biol Chem. 1997;272:21665–21668. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
