Modeling stochastic gene expression: implications for haploinsufficiency
- PMID: 9861023
- PMCID: PMC28097
- DOI: 10.1073/pnas.95.26.15641
Modeling stochastic gene expression: implications for haploinsufficiency
Abstract
There is increasing recognition that stochastic processes regulate highly predictable patterns of gene expression in developing organisms, but the implications of stochastic gene expression for understanding haploinsufficiency remain largely unexplored. We have used simulations of stochastic gene expression to illustrate that gene copy number and expression deactivation rates are important variables in achieving predictable outcomes. In gene expression systems with non-zero expression deactivation rates, diploid systems had a higher probability of uninterrupted gene expression than haploid systems and were more successful at maintaining gene product above a very low threshold. Systems with relatively rapid expression deactivation rates (unstable gene expression) had more predictable responses to a gradient of inducer than systems with slow or zero expression deactivation rates (stable gene expression), and diploid systems were more predictable than haploid, with or without dosage compensation. We suggest that null mutations of a single allele in a diploid organism could decrease the probability of gene expression and present the hypothesis that some haploinsufficiency syndromes might result from an increased susceptibility to stochastic delays of gene initiation or interruptions of gene expression.
Figures
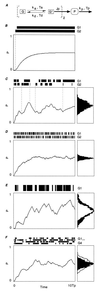
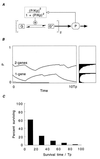
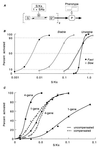
References
-
- Beaudet A L. In: Harrison’s Principles of Internal Medicine. Fauci A S, Braunwald E, Isselbacher K J, Wilson J D, Martin J B, Kasper D L, Hauser S L, Longo D L, editors. New York: McGraw–Hill; 1998. pp. 365–395.
-
- Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Genes Dev. 1990;4:1823–1834. - PubMed
-
- van Roon M A, Aten J A, van Oven C H, Charles R, Lamers W H. Dev Biol. 1989;136:508–516. - PubMed
-
- Dingemanse M A, de Boer P A, Moorman A F, Charles R, Lamers W H. Differentiation. 1994;56:153–162. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

