Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs
- PMID: 9861047
- PMCID: PMC28121
- DOI: 10.1073/pnas.95.26.15781
Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs
Abstract
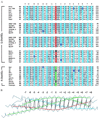
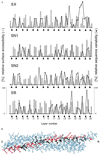
SNARE [soluble NSF (N-ethylmaleimide-sensitive fusion protein) attachment protein receptor] proteins are essential for membrane fusion and are conserved from yeast to humans. Sequence alignments of the most conserved regions were mapped onto the recently solved crystal structure of the heterotrimeric synaptic fusion complex. The association of the four alpha-helices in the synaptic fusion complex structure produces highly conserved layers of interacting amino acid side chains in the center of the four-helix bundle. Mutations in these layers reduce complex stability and cause defects in membrane traffic even in distantly related SNAREs. When syntaxin-4 is modeled into the synaptic fusion complex as a replacement of syntaxin-1A, no major steric clashes arise and the most variable amino acids localize to the outer surface of the complex. We conclude that the main structural features of the neuronal complex are highly conserved during evolution. On the basis of these features we have reclassified SNARE proteins into Q-SNAREs and R-SNAREs, and we propose that fusion-competent SNARE complexes generally consist of four-helix bundles composed of three Q-SNAREs and one R-SNARE.
Figures


References
-
- Ferro-Novick S, Jahn R. Nature (London) 1994;370:191–193. - PubMed
-
- Hanson P I, Heuser J E, Jahn R. Curr Opin Neurobiol. 1997;7:310–315. - PubMed
-
- Rothman J E. Nature (London) 1994;372:55–63. - PubMed
-
- Südhof T C. Nature (London) 1995;375:645–653. - PubMed
-
- Sutton R B, Fasshauer D, Jahn R, Brunger A T. Nature (London) 1998;395:347–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

