Two distinct mechanisms cause heterogeneity of 16S rRNA
- PMID: 9864315
- PMCID: PMC103534
- DOI: 10.1128/JB.181.1.78-82.1999
Two distinct mechanisms cause heterogeneity of 16S rRNA
Abstract
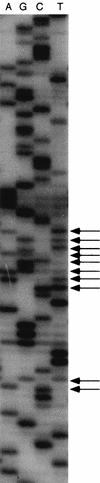
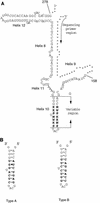
To investigate the frequency of heterogeneity among the multiple 16S rRNA genes within a single microorganism, we determined directly the 120-bp nucleotide sequences containing the hypervariable alpha region of the 16S rRNA gene from 475 Streptomyces strains. Display of the direct sequencing patterns revealed the existence of 136 heterogeneous loci among a total of 33 strains. The heterogeneous loci were detected only in the stem region designated helix 10. All of the substitutions conserved the relevant secondary structure. The 33 strains were divided into two groups: one group, including 22 strains, had less than two heterogeneous bases; the other group, including 11 strains, had five or more heterogeneous bases. The two groups were different in their combinations of heterogeneous bases. The former mainly contained transitional substitutions, and the latter was mainly composed of transversional substitutions, suggesting that at least two mechanisms, possibly misincorporation during DNA replication and horizontal gene transfer, cause rRNA heterogeneity.
Figures
Similar articles
-
Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes.Microbiology (Reading). 2000 Nov;146 ( Pt 11):2845-2854. doi: 10.1099/00221287-146-11-2845. Microbiology (Reading). 2000. PMID: 11065363
-
Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy.Microbiology (Reading). 2003 Jun;149(Pt 6):1493-1501. doi: 10.1099/mic.0.26132-0. Microbiology (Reading). 2003. PMID: 12777489
-
High intragenomic heterogeneity of 16S rRNA genes in a subset of Vibrio vulnificus strains from the western Mediterranean coast.Int Microbiol. 2010 Dec;13(4):179-88. doi: 10.2436/20.1501.01.124. Int Microbiol. 2010. PMID: 21404212
-
Comparison of partial 23S rDNA sequences from Rhizobium species.Can J Microbiol. 1997 Jun;43(6):526-33. doi: 10.1139/m97-075. Can J Microbiol. 1997. PMID: 9226872
-
Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria).PLoS One. 2017 Oct 26;12(10):e0186393. doi: 10.1371/journal.pone.0186393. eCollection 2017. PLoS One. 2017. PMID: 29073157 Free PMC article.
Cited by
-
Molecular Identification and Antimicrobial Potential of Streptomyces Species from Nepalese Soil.Int J Microbiol. 2020 Aug 27;2020:8817467. doi: 10.1155/2020/8817467. eCollection 2020. Int J Microbiol. 2020. PMID: 32908528 Free PMC article.
-
Application of multilocus sequence analysis (MLSA) for accurate identification of Legionella spp. Isolated from municipal fountains in Chengdu, China, based on 16S rRNA, mip, and rpoB genes.J Microbiol. 2012 Feb;50(1):127-36. doi: 10.1007/s12275-012-1243-1. Epub 2012 Feb 27. J Microbiol. 2012. PMID: 22367947
-
Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases.Clin Microbiol Rev. 2004 Oct;17(4):840-62, table of contents. doi: 10.1128/CMR.17.4.840-862.2004. Clin Microbiol Rev. 2004. PMID: 15489351 Free PMC article. Review.
-
Rates and consequences of recombination between rRNA operons.J Bacteriol. 2003 Feb;185(3):966-72. doi: 10.1128/JB.185.3.966-972.2003. J Bacteriol. 2003. PMID: 12533472 Free PMC article.
-
Ribosomal multi-operon diversity: an original perspective on the genus Aeromonas.PLoS One. 2012;7(9):e46268. doi: 10.1371/journal.pone.0046268. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23032081 Free PMC article.
References
-
- Amabile-Cuevas C F, Chicurel M E. Bacterial plasmids and gene flux. Cell. 1992;70:189–199. - PubMed
-
- Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. - PubMed
-
- Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

