Two distinct mechanisms cause heterogeneity of 16S rRNA
- PMID: 9864315
- PMCID: PMC103534
- DOI: 10.1128/JB.181.1.78-82.1999
Two distinct mechanisms cause heterogeneity of 16S rRNA
Abstract
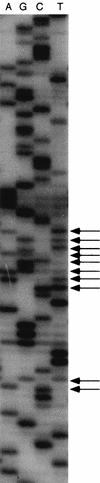
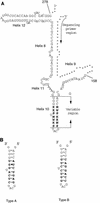
To investigate the frequency of heterogeneity among the multiple 16S rRNA genes within a single microorganism, we determined directly the 120-bp nucleotide sequences containing the hypervariable alpha region of the 16S rRNA gene from 475 Streptomyces strains. Display of the direct sequencing patterns revealed the existence of 136 heterogeneous loci among a total of 33 strains. The heterogeneous loci were detected only in the stem region designated helix 10. All of the substitutions conserved the relevant secondary structure. The 33 strains were divided into two groups: one group, including 22 strains, had less than two heterogeneous bases; the other group, including 11 strains, had five or more heterogeneous bases. The two groups were different in their combinations of heterogeneous bases. The former mainly contained transitional substitutions, and the latter was mainly composed of transversional substitutions, suggesting that at least two mechanisms, possibly misincorporation during DNA replication and horizontal gene transfer, cause rRNA heterogeneity.
Figures
References
-
- Amabile-Cuevas C F, Chicurel M E. Bacterial plasmids and gene flux. Cell. 1992;70:189–199. - PubMed
-
- Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. - PubMed
-
- Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

