Identification and properties of the genes encoding microcin E492 and its immunity protein
- PMID: 9864332
- PMCID: PMC103551
- DOI: 10.1128/JB.181.1.212-217.1999
Identification and properties of the genes encoding microcin E492 and its immunity protein
Abstract
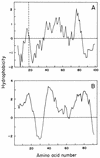
The gene coding for the immunity protein (mceB) and the structural gene of microcin E492 (mceA), a low-molecular-weight channel-forming bacteriocin produced by a strain of Klebsiella pneumoniae, have been characterized. The microcin gene codes for a precursor protein of either 99 or 103 amino acids. Protein sequencing of the N-terminal region of microcin E492 unequivocally identified this gene as the microcin structural gene and indicated that this microcin is synthesized as a precursor protein that is cleaved at either amino acid 15 or 19, at a site resembling the double-glycine motif. The gene encoding the 95-amino-acid immunity protein (mceB) was identified by cloning the DNA segment that encodes only this polypeptide into an expression vector and demonstrating the acquisition of immunity to microcin E492. As expected, the immunity protein was found to be associated with the inner membrane. Analysis of the DNA sequence indicates that these genes belong to the same family as microcin 24, and they do not share structural motifs with any other known channel-forming bacteriocin. The organization of the microcin- and immunity protein-encoding genes suggests that they are coordinately expressed.
Figures



References
-
- Abee T. Pore-forming bacteriocins of Gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates and John Wiley & Sons, Inc.; 1992.
-
- Baba T, Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. - PubMed
-
- Baeza, M., C. Hetz, C. Villota, J. E. Villanueva, and R. Lagos. Unpublished data.
-
- Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

