Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis
- PMID: 9864354
- PMCID: PMC2175213
- DOI: 10.1083/jcb.143.7.1775
Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis
Abstract
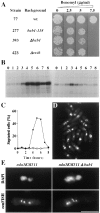
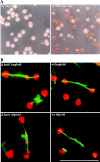
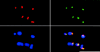
The spindle checkpoint ensures proper chromosome segregation by delaying anaphase until all chromosomes are correctly attached to the mitotic spindle. We investigated the role of the fission yeast bub1 gene in spindle checkpoint function and in unperturbed mitoses. We find that bub1(+) is essential for the fission yeast spindle checkpoint response to spindle damage and to defects in centromere function. Activation of the checkpoint results in the recruitment of Bub1 to centromeres and a delay in the completion of mitosis. We show that Bub1 also has a crucial role in normal, unperturbed mitoses. Loss of bub1 function causes chromosomes to lag on the anaphase spindle and an increased frequency of chromosome loss. Such genomic instability is even more dramatic in Deltabub1 diploids, leading to massive chromosome missegregation events and loss of the diploid state, demonstrating that bub1(+ )function is essential to maintain correct ploidy through mitosis. As in larger eukaryotes, Bub1 is recruited to kinetochores during the early stages of mitosis. However, unlike its vertebrate counterpart, a pool of Bub1 remains centromere-associated at metaphase and even until telophase. We discuss the possibility of a role for the Bub1 kinase after the metaphase-anaphase transition.
Figures




References
-
- Allshire RC. Centromeres, checkpoints and chromatid cohesion. Curr Opin Genet Dev. 1997;7:264–273. - PubMed
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat J-P, Granston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
-
- Allshire RC, Javerzat J-P, Redhead NJ, Granston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Altshul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

