The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors
- PMID: 9870940
- PMCID: PMC6782383
- DOI: 10.1523/JNEUROSCI.19-01-00072.1999
The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors
Abstract
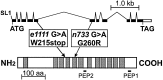
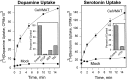
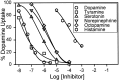
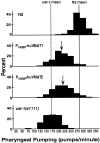
We have identified the Caenorhabditis elegans homolog of the mammalian vesicular monoamine transporters (VMATs); it is 47% identical to human VMAT1 and 49% identical to human VMAT2. C. elegans VMAT is associated with synaptic vesicles in approximately 25 neurons, including all of the cells reported to contain dopamine and serotonin, plus a few others. When C. elegans VMAT is expressed in mammalian cells, it has serotonin and dopamine transport activity; norepinephrine, tyramine, octopamine, and histamine also have high affinity for the transporter. The pharmacological profile of C. elegans VMAT is closer to mammalian VMAT2 than VMAT1. The C. elegans VMAT gene is cat-1; cat-1 knock-outs are totally deficient for VMAT immunostaining and for dopamine-mediated sensory behaviors, yet they are viable and grow relatively well. The cat-1 mutant phenotypes can be rescued by C. elegans VMAT constructs and also (at least partially) by human VMAT1 or VMAT2 transgenes. It therefore appears that the function of amine neurotransmitters can be completely dependent on their loading into synaptic vesicles.
Figures






References
-
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. - PubMed
-
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. - PubMed
-
- Alfonso A, Grundahl K, Duerr JS, Han H-P, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science. 1993;261:617–619. - PubMed
-
- Avery L, Horvitz HR. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J Exp Zool. 1990;253:263–270. - PubMed
-
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991a;7:729–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
