Enumeration of marine viruses in culture and natural samples by flow cytometry
- PMID: 9872758
- PMCID: PMC90981
- DOI: 10.1128/AEM.65.1.45-52.1999
Enumeration of marine viruses in culture and natural samples by flow cytometry
Abstract
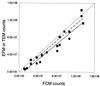
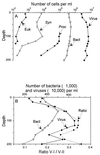
Flow cytometry (FCM) was successfully used to enumerate viruses in seawater after staining with the nucleic acid-specific dye SYBR Green-I. The technique was first optimized by using the Phaeocystis lytic virus PpV-01. Then it was used to analyze natural samples from different oceanic locations. Virus samples were fixed with 0.5% glutaraldehyde and deep frozen for delayed analysis. The samples were then diluted in Tris-EDTA buffer and analyzed in the presence of SYBR Green-I. A duplicate sample was heated at 80 degreesC in the presence of detergent before analysis. Virus counts obtained by FCM were highly correlated to, although slightly higher than, those obtained by epifluorescence microscopy or by transmission electron microscopy (r = 0.937, n = 14, and r = 0.96, n = 8, respectively). Analysis of a depth profile from the Mediterranean Sea revealed that the abundance of viruses displayed the same vertical trend as that of planktonic cells. FCM permits us to distinguish between at least two and sometimes three virus populations in natural samples. Because of its speed and accuracy, FCM should prove very useful for studies of virus infection in cultures and should allow us to better understand the structure and dynamics of virus populations in natural waters.
Figures




References
-
- Bergh Ø, Børsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. - PubMed
-
- Bratbak G, Heldal M. Total count of viruses in aquatic environments. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Current methods in aquatic microbial ecology. London, England: Lewis Publishers; 1993. pp. 135–138.
-
- Bratbak G, Egge J K, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48.
LinkOut - more resources
Full Text Sources
Other Literature Sources

