Pleiotropic effects of adaptation to a single carbon source for growth on alternative substrates
- PMID: 9872788
- PMCID: PMC91011
- DOI: 10.1128/AEM.65.1.264-269.1999
Pleiotropic effects of adaptation to a single carbon source for growth on alternative substrates
Abstract
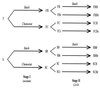
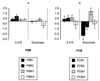
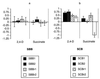
It is frequently assumed that populations of genetically modified microorganisms will perform their intended function and then disappear from the environment due to inherent fitness disadvantages resulting from their genetic alteration. However, modified organisms used in bioremediation can be expected to adapt evolutionarily to growth on the anthropogenic substrate that they are intended to degrade. If such adaptation results in improved competitiveness for alternative, naturally occurring substrates, then this will increase the likelihood that the modified organisms will persist in the environment. In this study, bacteria capable of degrading the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) were used to test the effects of evolutionary adaptation to one substrate on fitness during growth on an alternative substrate. Twenty lineages of bacteria were allowed to evolve under abundant resource conditions on either 2,4-D or succinate as their sole carbon source. The competitiveness of each evolved line was then measured relative to that of its ancestor for growth on both substrates. Only three derived lines showed a clear drop in fitness on the alternative substrate after demonstrable adaptation to their selective substrate, while five derived lines showed significant simultaneous increases in fitness on both their selective and alternative substrates. These data demonstrate that adaptation to an anthropogenic substrate can pleiotropically increase competitiveness for an alternative natural substrate and therefore increase the likelihood that a genetically modified organism will persist in the environment.
Figures

Similar articles
-
Effect of integration of a GFP reporter gene on fitness of Ralstonia eutropha during growth with 2,4-dichlorophenoxyacetic acid.Environ Microbiol. 2003 Oct;5(10):878-87. doi: 10.1046/j.1462-2920.2003.00479.x. Environ Microbiol. 2003. PMID: 14510841
-
Asymmetric, bimodal trade-offs during adaptation of Methylobacterium to distinct growth substrates.Evolution. 2009 Nov;63(11):2816-30. doi: 10.1111/j.1558-5646.2009.00757.x. Epub 2009 Jun 22. Evolution. 2009. PMID: 19545267
-
Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies.Appl Environ Microbiol. 2017 Jun 16;83(13):e00410-17. doi: 10.1128/AEM.00410-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28455337 Free PMC article.
-
Evaluating the fate of genetically modified microorganisms in the environment: are they inherently less fit?Experientia. 1993 Mar 15;49(3):201-9. doi: 10.1007/BF01923527. Experientia. 1993. PMID: 8458406 Review.
-
Safety and nutritional assessment of GM plants and derived food and feed: the role of animal feeding trials.Food Chem Toxicol. 2008 Mar;46 Suppl 1:S2-70. doi: 10.1016/j.fct.2008.02.008. Epub 2008 Feb 13. Food Chem Toxicol. 2008. PMID: 18328408 Review.
Cited by
-
Predatory Bacteria Select for Sustained Prey Diversity.Microorganisms. 2021 Oct 2;9(10):2079. doi: 10.3390/microorganisms9102079. Microorganisms. 2021. PMID: 34683400 Free PMC article.
-
Indirect evolution of social fitness inequalities and facultative social exploitation.Proc Biol Sci. 2018 Mar 28;285(1875):20180054. doi: 10.1098/rspb.2018.0054. Proc Biol Sci. 2018. PMID: 29593113 Free PMC article.
-
Stress-induced evolution and the biosafety of genetically modified microorganisms released into the environment.J Biosci. 2001 Dec;26(5):667-83. doi: 10.1007/BF02704764. J Biosci. 2001. PMID: 11807296 Review.
-
Aneuploidy Enables Cross-Adaptation to Unrelated Drugs.Mol Biol Evol. 2019 Aug 1;36(8):1768-1782. doi: 10.1093/molbev/msz104. Mol Biol Evol. 2019. PMID: 31028698 Free PMC article.
-
How does resource supply affect evolutionary diversification?Proc Biol Sci. 2007 Jan 7;274(1606):73-8. doi: 10.1098/rspb.2006.3703. Proc Biol Sci. 2007. PMID: 17015335 Free PMC article.
References
-
- Andrews K J, Hegeman G D. Selective disadvantage of nonfunctional protein synthesis in Escherichia coli. J Mol Evol. 1976;8:317–328. - PubMed
-
- Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli. Mol Gen Genet. 1994;242:595–601. - PubMed
-
- Bouma J E, Lenski R E. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. - PubMed
-
- Boyle J J, Shann J R. Biodegradation of pheno, 2,4-DCP, 2,4-D, and 2,4,5-T in field collected rhizosphere and nonrhizosphere soils. J Environ Qual. 1995;24:782–785.
LinkOut - more resources
Full Text Sources
Other Literature Sources

