A microfabricated device for sizing and sorting DNA molecules
- PMID: 9874762
- PMCID: PMC15083
- DOI: 10.1073/pnas.96.1.11
A microfabricated device for sizing and sorting DNA molecules
Abstract
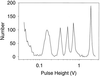
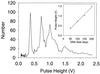
We have demonstrated a microfabricated single-molecule DNA sizing device. This device does not depend on mobility to measure molecule size, is 100 times faster than pulsed-field gel electrophoresis, and has a resolution that improves with increasing DNA length. It also requires a million times less sample than pulsed-field gel electrophoresis and has comparable resolution for large molecules. Here we describe the fabrication and use of the single-molecule DNA sizing device for sizing and sorting DNA restriction digests and ladders spanning 2-200 kbp.
Figures


Similar articles
-
Scrambling of bands in gel electrophoresis of DNA.Nucleic Acids Res. 1988 Jun 24;16(12):5427-37. doi: 10.1093/nar/16.12.5427. Nucleic Acids Res. 1988. PMID: 2838816 Free PMC article.
-
Pulsed field capillary electrophoresis of multikilobase length nucleic acids in dilute methyl cellulose solutions.Anal Chem. 1994 Oct 1;66(19):3081-5. doi: 10.1021/ac00091a015. Anal Chem. 1994. PMID: 7978303
-
Enhanced discrimination of DNA molecules in nanofluidic channels through multiple measurements.Lab Chip. 2012 Mar 21;12(6):1094-101. doi: 10.1039/c2lc20771k. Epub 2012 Feb 2. Lab Chip. 2012. PMID: 22298224 Free PMC article.
-
Electrophoretic mobility of lambda phage HIND III and HAE III DNA fragments in agarose gels: a detailed study.Biopolymers. 1987 May;26(5):727-42. doi: 10.1002/bip.360260512. Biopolymers. 1987. PMID: 3036265 No abstract available.
-
Integrated CMOS microchip system with capillary array electrophoresis.Anal Bioanal Chem. 2002 Jul;373(6):399-403. doi: 10.1007/s00216-002-1353-4. Epub 2002 Jun 29. Anal Bioanal Chem. 2002. PMID: 12172674
Cited by
-
Single molecule analysis of bacterial polymerase chain reaction products in submicrometer fluidic channels.Biomicrofluidics. 2007 Sep 20;1(3):34105. doi: 10.1063/1.2789565. Biomicrofluidics. 2007. PMID: 19693361 Free PMC article.
-
Conformation, length, and speed measurements of electrodynamically stretched DNA in nanochannels.Biophys J. 2008 Jul;95(1):273-86. doi: 10.1529/biophysj.107.121020. Epub 2008 Mar 13. Biophys J. 2008. PMID: 18339746 Free PMC article.
-
Single-file electrophoretic transport and counting of individual DNA molecules in surfactant nanotubes.Proc Natl Acad Sci U S A. 2005 Jun 28;102(26):9127-32. doi: 10.1073/pnas.0500081102. Epub 2005 Jun 16. Proc Natl Acad Sci U S A. 2005. PMID: 15961544 Free PMC article.
-
Fabrication Methods for Microfluidic Devices: An Overview.Micromachines (Basel). 2021 Mar 18;12(3):319. doi: 10.3390/mi12030319. Micromachines (Basel). 2021. PMID: 33803689 Free PMC article. Review.
-
A Visualization Technique of a Unique pH Distribution around an Ion Depletion Zone in a Microchannel by Using a Dual-Excitation Ratiometric Method.Micromachines (Basel). 2018 Apr 2;9(4):167. doi: 10.3390/mi9040167. Micromachines (Basel). 2018. PMID: 30424100 Free PMC article.
References
-
- Burmeister M, Ulanovsky L, editors. Pulsed-Field Gel Electrophoresis: Protocols, Methods, and Theories. Totowa, NJ: Humana; 1992.
-
- Haugland R P. Handbook of Fluorescent Probes and Research Chemicals. Eugene, OR: Molecular Probes; 1996.
-
- Castro A, Fairfield F R, Schera E B. Anal Chem. 1993;65:849–852.
-
- Guo X H, Huff E J, Schwartz D C. Nature (London) 1992;359:783–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

