NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas
- PMID: 9874798
- PMCID: PMC15119
- DOI: 10.1073/pnas.96.1.214
NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas
Abstract
Using differential display PCR, we have identified a gene [NOEY2, ARHI (designation by the Human Gene Nomenclature Committee)] with high homology to ras and rap that is expressed consistently in normal ovarian and breast epithelial cells but not in ovarian and breast cancers. Reexpression of NOEY2 through transfection suppresses clonogenic growth of breast and ovarian cancer cells. Growth suppression was associated with down-regulation of the cyclin D1 promoter activity and induction of p21(WAF1/CIP1). In an effort to identify mechanisms leading to NOEY2 silencing in cancer, we found that the gene is expressed monoallelically and is imprinted maternally. Loss of heterozygosity of the gene was detected in 41% of ovarian and breast cancers. In most of cancer samples with loss of heterozygosity, the nonimprinted functional allele was deleted. Thus, NOEY2 appears to be a putative imprinted tumor suppressor gene whose function is abrogated in ovarian and breast cancers.
Figures

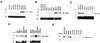



References
-
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. - PubMed
-
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Nature (London) 1995;378:789–792. - PubMed
-
- Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, et al. Science. 1990;250:1233–1238. - PubMed
-
- Nagai H, Negrini M, Carter S L, Gillum D R, Rosenberg A L, Schwartz G F, Croce C M. Cancer Res. 1995;55:1752–1757. - PubMed
-
- Loupart M L, Armour J, Walker R, Adams S, Brammar W, Varley J. Genes Chromosomes Cancer. 1995;12:16–23. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

