The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family
- PMID: 9874811
- PMCID: PMC15132
- DOI: 10.1073/pnas.96.1.290
The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family
Abstract
The ability of the shoot apical meristem to multiply and distribute its meristematic potential through the formation of axillary meristems is essential for the diversity of forms and growth habits of higher plants. In the lateral suppressor mutant of tomato the initiation of axillary meristems is prevented, thus offering the unique opportunity to study the molecular mechanisms underlying this important function of the shoot apical meristem. We report here the isolation of the Lateral suppressor gene by positional cloning and show that the mutant phenotype is caused by a complete loss of function of a new member of the VHIID family of plant regulatory proteins.
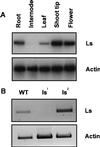
Figures



Similar articles
-
The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems.Proc Natl Acad Sci U S A. 2002 Jan 22;99(2):1064-9. doi: 10.1073/pnas.022516199. Proc Natl Acad Sci U S A. 2002. PMID: 11805344 Free PMC article.
-
Ectopic expression a tomato KNOX Gene Tkn4 affects the formation and the differentiation of meristems and vasculature.Plant Mol Biol. 2015 Dec;89(6):589-605. doi: 10.1007/s11103-015-0387-x. Epub 2015 Oct 11. Plant Mol Biol. 2015. PMID: 26456092
-
Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis.Plant Cell. 2006 Mar;18(3):586-97. doi: 10.1105/tpc.105.038745. Epub 2006 Feb 3. Plant Cell. 2006. PMID: 16461581 Free PMC article.
-
Genetic control of branching in Arabidopsis and tomato.Curr Opin Plant Biol. 1999 Feb;2(1):51-5. doi: 10.1016/s1369-5266(99)80010-7. Curr Opin Plant Biol. 1999. PMID: 10047573 Review.
-
Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato.J Exp Bot. 2006;57(13):3405-14. doi: 10.1093/jxb/erl106. Epub 2006 Sep 27. J Exp Bot. 2006. PMID: 17005925 Review.
Cited by
-
Genome-Wide Identification, Evolutionary Analysis, and Stress Responses of the GRAS Gene Family in Castor Beans.Int J Mol Sci. 2016 Jun 24;17(7):1004. doi: 10.3390/ijms17071004. Int J Mol Sci. 2016. PMID: 27347937 Free PMC article.
-
Multifaceted roles of GRAS transcription factors in growth and stress responses in plants.iScience. 2022 Aug 28;25(9):105026. doi: 10.1016/j.isci.2022.105026. eCollection 2022 Sep 16. iScience. 2022. PMID: 36117995 Free PMC article. Review.
-
Evolution and functional analysis of the GRAS family genes in six Rosaceae species.BMC Plant Biol. 2022 Dec 6;22(1):569. doi: 10.1186/s12870-022-03925-x. BMC Plant Biol. 2022. PMID: 36471247 Free PMC article.
-
Loss of Lateral suppressor gene is associated with evolution of root nodule symbiosis in Leguminosae.Genome Biol. 2024 Sep 30;25(1):250. doi: 10.1186/s13059-024-03393-6. Genome Biol. 2024. PMID: 39350172 Free PMC article.
-
Gibberellin signaling: biosynthesis, catabolism, and response pathways.Plant Cell. 2002;14 Suppl(Suppl):S61-80. doi: 10.1105/tpc.010476. Plant Cell. 2002. PMID: 12045270 Free PMC article. Review. No abstract available.
References
-
- Steeves T A, Sussex I M. Patterns in Plant Development. 2nd Ed. Cambridge, U.K.: Cambridge Univ. Press; 1989.
-
- Cline M G. Am J Bot. 1997;84:1064–1069. - PubMed
-
- Napoli C A, Ruehle J. J Hered. 1996;87:371–377.
-
- Malayer J C, Guard A T. Am J Bot. 1964;51:140–143.
-
- Williams W. Heredity. 1960;14:285–296.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

