The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling
- PMID: 9880329
- PMCID: PMC25156
- DOI: 10.1091/mbc.10.1.91
The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling
Abstract
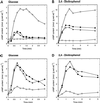
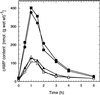
The yeast Saccharomyces cerevisiae contains two genes, PDE1 and PDE2, which respectively encode a low-affinity and a high-affinity cAMP phosphodiesterase. The physiological function of the low-affinity enzyme Pde1 is unclear. We show that deletion of PDE1, but not PDE2, results in a much higher cAMP accumulation upon addition of glucose or upon intracellular acidification. Overexpression of PDE1, but not PDE2, abolished the agonist-induced cAMP increases. These results indicate a specific role for Pde1 in controlling glucose and intracellular acidification-induced cAMP signaling. Elimination of a putative protein kinase A (PKA) phosphorylation site by mutagenesis of serine252 into alanine resulted in a Pde1(ala252) allele that apparently had reduced activity in vivo. Its presence in a wild-type strain partially enhanced the agonist-induced cAMP increases compared with pde1Delta. The difference between the Pde1(ala252) allele and wild-type Pde1 was strongly dependent on PKA activity. In a RAS2(val19) pde2Delta background, the Pde1(ala252) allele caused nearly the same hyperaccumulation of cAMP as pde1Delta, while its expression in a PKA-attenuated strain caused the same reduction in cAMP hyperaccumulation as wild-type Pde1. These results suggest that serine252 might be the first target site for feedback inhibition of cAMP accumulation by PKA. We show that Pde1 is rapidly phosphorylated in vivo upon addition of glucose to glycerol-grown cells, and this activation is absent in the Pde1(ala252) mutant. Pde1 belongs to a separate class of phosphodiesterases and is the first member shown to be phosphorylated. However, in vitro the Pde1(ala252) enzyme had the same catalytic activity as wild-type Pde1, both in crude extracts and after extensive purification. This indicates that the effects of the S252A mutation are not caused by simple inactivation of the enzyme. In vitro phosphorylation of Pde1 resulted in a modest and variable increase in activity, but only in crude extracts. This was absent in Pde1(ala252), and phosphate incorporation was strongly reduced. Apparently, phosphorylation of Pde1 does not change its intrinsic activity or affinity for cAMP but appears to be important in vivo for protein-protein interaction or for targeting Pde1 to a specific subcellular location. The PKA recognition site is conserved in the corresponding region of the Schizosaccharomyces pombe and Candida albicans Pde1 homologues, possibly indicating a similar control by phosphorylation.
Figures




References
-
- Argüelles JC, Mbonyi K, Van Aelst L, Vanhalewyn M, Jans AWH, Thevelein JM. Absence of glucose-induced cAMP signaling in the Saccharomyces cerevisiae mutants cat1 and cat3 which are deficient in derepression of glucose-repressible proteins. Arch Microbiol. 1990;154:199–205. - PubMed
-
- Beullens M, Mbonyi K, Geerts L, Gladines D, Detremerie K, Jans AWH, Thevelein JM. Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1988;172:227–231. - PubMed
-
- Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5′-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
-
- Broach JR, Deschenes RJ. The function of RAS genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. - PubMed
-
- Broek D, Samiy N, Fasano O, Fujiyama A, Tamanoi F, Northup J, Wigler M. Differential activation of yeast adenylate cyclase by wild type and mutant Ras proteins. Cell. 1985;41:763–769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

