Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis
- PMID: 9880351
- PMCID: PMC32209
- DOI: 10.1104/pp.119.1.101
Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis
Abstract
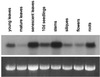
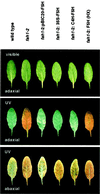
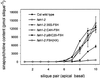
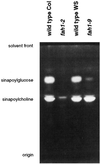
Sinapic acid is an intermediate in syringyl lignin biosynthesis in angiosperms, and in some taxa serves as a precursor for soluble secondary metabolites. The biosynthesis and accumulation of the sinapate esters sinapoylglucose, sinapoylmalate, and sinapoylcholine are developmentally regulated in Arabidopsis and other members of the Brassicaceae. The FAH1 locus of Arabidopsis encodes the enzyme ferulate-5-hydroxylase (F5H), which catalyzes the rate-limiting step in syringyl lignin biosynthesis and is required for the production of sinapate esters. Here we show that F5H expression parallels sinapate ester accumulation in developing siliques and seedlings, but is not rate limiting for their biosynthesis. RNA gel-blot analysis indicated that the tissue-specific and developmentally regulated expression of F5H mRNA is distinct from that of other phenylpropanoid genes. Efforts to identify constructs capable of complementing the sinapate ester-deficient phenotype of fah1 mutants demonstrated that F5H expression in leaves is dependent on sequences 3' of the F5H coding region. In contrast, the positive regulatory function of the downstream region is not required for F5H transcript or sinapoylcholine accumulation in embryos.
Figures




References
-
- An G. Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol. 1987;153:292–305.
-
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. - PubMed
-
- Bouchereau A, Hamelin J, Lamour I, Renard M, Larher F. Distribution of sinapine and related compounds in seeds of Brassica and allied genera. Phytochemistry. 1991;30:1873–1881.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

