Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize
- PMID: 9880368
- PMCID: PMC32228
- DOI: 10.1104/pp.119.1.255
Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize
Abstract
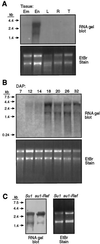
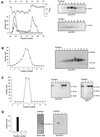
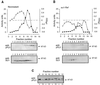
This study identified and purified specific isoamylase- and pullulanase-type starch-debranching enzymes (DBEs) present in developing maize (Zea mays L.) endosperm. The cDNA clone Zpu1 was isolated based on its homology with a rice (Oryza sativa L.) cDNA coding for a pullulanase-type DBE. Comparison of the protein product, ZPU1, with 18 other DBEs identified motifs common to both isoamylase- and pullulanase-type enzymes, as well as class-specific sequence blocks. Hybridization of Zpu1 to genomic DNA defined a single-copy gene, zpu1, located on chromosome 2. Zpu1 mRNA was abundant in endosperm throughout starch biosynthesis, but was not detected in the leaf or the root. Anti-ZPU1 antiserum specifically recognized the approximately 100-kD ZPU1 protein in developing endosperm, but not in leaves. Pullulanase- and isoamylase-type DBEs were purified from extracts of developing maize kernels. The pullulanase-type activity was identified as ZPU1 and the isoamylase-type activity as SU1. Mutations of the sugary1 (su1) gene are known to cause deficiencies of SU1 isoamylase and a pullulanase-type DBE. ZPU1 activity, protein level, and electrophoretic mobility were altered in su1-mutant kernels, indicating that it is the affected pullulanase-type DBE. The Zpu1 transcript levels were equivalent in nonmutant and su1-mutant kernels, suggesting that coordinated regulation of ZPU1 and SU1 occurs posttranscriptionally.
Figures



References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989.
-
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. - PubMed
-
- Beavis W, Berlyn M, Burr B, Chandler V, Coe E, Fauron C, Nelson O, Polacco M, Rodermel S, Sachs M and others. A standard for maize genetics nomenclature. Maize Genet Coop Newsl. 1995;69:182–184.
-
- Burr B, Burr FA, Matz EC. Maize molecular map (Zea mays) 2n=20. In: O'Brien SJ, editor. Genetic Maps. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 190–203.
-
- Burr B, Burr FA, Matz EC. Mapping genes with recombinant inbreds. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 249–254.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

