Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences
- PMID: 9882302
- PMCID: PMC103921
- DOI: 10.1128/JVI.73.2.1010-1022.1999
Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences
Abstract
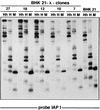
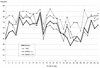
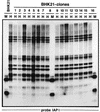
The insertion of adenovirus type 12 (Ad12) DNA into the hamster genome and the transformation of these cells by Ad12 can lead to marked alterations in the levels of DNA methylation in several cellular genes and DNA segments. Since such alterations in DNA methylation patterns are likely to affect the transcription patterns of cellular genes, it is conceivable that these changes have played a role in the generation or the maintenance of the Ad12-transformed phenotype. We have now isolated clonal BHK21 hamster cell lines that carry in their genomes bacteriophage lambda and plasmid pSV2neo DNAs in an integrated state. Most of these cell lines contain one or multiple copies of integrated lambda DNA, which often colocalize with the pSV2neo DNA, usually in a single chromosomal site as determined by the fluorescent in situ hybridization technique. In different cell lines, the loci of foreign DNA insertion are different. The inserted bacteriophage lambda DNA frequently becomes de novo methylated. In some of the thus-generated hamster cell lines, the levels of DNA methylation in the retrotransposon genomes of the endogenous intracisternal A particles (IAP) are increased in comparison to those in the non-lambda-DNA-transgenic BHK21 cell lines. These changes in the methylation patterns of the IAP subclone I (IAPI) segment have been documented by restriction analyses with methylation-sensitive restriction endonucleases followed by Southern transfer hybridization and phosphorimager quantitation. The results of genomic sequencing experiments using the bisulfite protocol yielded additional evidence for alterations in the patterns of DNA methylation in selected segments of the IAPI sequences. In these experiments, the nucleotide sequences in >330 PCR-generated cloned DNA molecules were determined. Upon prolonged cultivation of cell lines with altered cellular methylation patterns, these differences became less apparent, perhaps due to counterselection of the transgenic cells. The possibility existed that the hamster BHK21 cell genomes represent mosaics with respect to DNA methylation in the IAPI segment. Hence, some of the cells with the patterns observed after lambda DNA integration might have existed prior to lambda DNA integration and been selected by chance. A total of 66 individual BHK21 cell clones from the BHK21 cell stock have been recloned up to three times, and the DNAs of these cell populations have been analyzed for differences in IAPI methylation patterns. None have been found. These patterns are identical among the individual BHK21 cell clones and identical to the patterns of the originally used BHK21 cell line. Similar results have been obtained with nine clones isolated from BHK21 cells mock transfected by the Ca2+-phosphate precipitation procedure with DNA omitted from the transfection mixture. In four clonal sublines of nontransgenic control BHK21 cells, genomic sequencing of 335 PCR-generated clones by the bisulfite protocol revealed 5'-CG-3' methylation levels in the IAPI segment that were comparable to those in the uncloned BHK21 cell line. We conclude that the observed changes in the DNA methylation patterns in BHK21 cells with integrated lambda DNA are unlikely to preexist or to be caused by the transfection procedure. Our data support the interpretation that the insertion of foreign DNA into a preexisting mammalian genome can alter the cellular patterns of DNA methylation, perhaps via changes in chromatin structure. The cellular sites affected by and the extent of these changes could depend on the site and size of foreign DNA insertion.
Figures





Similar articles
-
Chromosomal insertion of foreign (adenovirus type 12, plasmid, or bacteriophage lambda) DNA is associated with enhanced methylation of cellular DNA segments.Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5515-9. doi: 10.1073/pnas.92.12.5515. Proc Natl Acad Sci U S A. 1995. PMID: 7777540 Free PMC article.
-
Foreign DNA integration. Genome-wide perturbations of methylation and transcription in the recipient genomes.J Biol Chem. 2001 Apr 27;276(17):14271-8. doi: 10.1074/jbc.M009380200. Epub 2001 Jan 18. J Biol Chem. 2001. PMID: 11278495
-
The initiation of de novo methylation of foreign DNA integrated into a mammalian genome is not exclusively targeted by nucleotide sequence.J Virol. 1995 Feb;69(2):1226-42. doi: 10.1128/JVI.69.2.1226-1242.1995. J Virol. 1995. PMID: 7815498 Free PMC article.
-
A new concept in (adenoviral) oncogenesis: integration of foreign DNA and its consequences.Biochim Biophys Acta. 1996 Oct 9;1288(2):F79-99. doi: 10.1016/0304-419x(96)00024-8. Biochim Biophys Acta. 1996. PMID: 8876634 Review.
-
Foreign DNA integration--perturbations of the genome--oncogenesis.Ann N Y Acad Sci. 2001 Sep;945:276-88. doi: 10.1111/j.1749-6632.2001.tb03896.x. Ann N Y Acad Sci. 2001. PMID: 11708490 Review.
Cited by
-
Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia.Plant Mol Biol. 2004 Mar;54(4):571-82. doi: 10.1023/B:PLAN.0000038270.48326.7a. Plant Mol Biol. 2004. PMID: 15316290
-
Relationships between DNA methylation and expression in erythrocyte membrane protein (band 3, protein 4.2, and beta-spectrin) genes during human erythroid development and differentiation.Int J Hematol. 2005 Dec;82(5):422-9. doi: 10.1532/IJH97.05058. Int J Hematol. 2005. PMID: 16533746
-
Inheritable epigenetic response towards foreign DNA entry by mammalian host cells: a guardian of genomic stability.Epigenetics. 2018;13(12):1141-1153. doi: 10.1080/15592294.2018.1549463. Epub 2018 Dec 12. Epigenetics. 2018. PMID: 30458693 Free PMC article. Review.
-
Molecular mechanism of immune response induced by foreign plasmid DNA after oral administration in mice.World J Gastroenterol. 2007 Jul 28;13(28):3847-54. doi: 10.3748/wjg.v13.i28.3847. World J Gastroenterol. 2007. PMID: 17657840 Free PMC article.
-
Methylated Host Cell Gene Promoters and Human Papillomavirus Type 16 and 18 Predicting Cervical Lesions and Cancer.PLoS One. 2015 Jun 9;10(6):e0129452. doi: 10.1371/journal.pone.0129452. eCollection 2015. PLoS One. 2015. PMID: 26057381 Free PMC article.
References
-
- Achten S, Behn-Krappa A, Jücker M, Sprengel J, Hölker I, Schmitz B, Tesch H, Diehl V, Doerfler W. Patterns of DNA methylation in selected human genes in different Hodgkin’s lymphoma and leukemia cell lines and in normal human lymphocytes. Cancer Res. 1991;51:3702–3709. - PubMed
-
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium isothiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
-
- Doerfler W. DNA methylation—a regulatory signal in eukaryotic gene expression. J Gen Virol. 1981;57:1–20. - PubMed
-
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. - PubMed
-
- Doerfler W, Gahlmann R, Stabel S, Deuring R, Lichtenberg U, Schulz M, Eick D, Leisten R. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr Top Microbiol Immunol. 1983;109:193–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

