Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses
- PMID: 9882304
- PMCID: PMC103923
- DOI: 10.1128/JVI.73.2.1036-1045.1999
Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses
Abstract
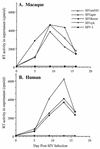
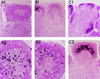
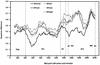
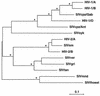
The human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2) appear to have originated by cross-species transmission of simian immunodeficiency virus (SIV) from asymptomatically infected African primates. Few of the SIVs characterized to date efficiently infect human primary lymphocytes. Interesting, two of the three identified to infect such cultures (SIVsm and SIVcpz) have appeared in human populations as genetically related HIVs. In the present study, we characterized a novel SIV isolate from an East African monkey of the Cercopithecus genus, the l'hoest monkey (C. l'hoesti), which we designated SIVlhoest. This SIV isolate efficiently infected both human and macaque lymphocytes and resulted in a persistent infection of macaques, characterized by high primary virus load and a progressive decline in circulating CD4 lymphocytes, consistent with progression to AIDS. Phylogenetic analyses showed that SIVlhoest is genetically distinct from other previously characterized primate lentiviruses but clusters in the same major lineage as SIV from mandrills (SIVmnd), a West African primate species. Given the geographic distance between the ranges of l'hoest monkeys and mandrills, this may indicate that SIVmnd arose through cross-species transmission from close relatives of l'hoest monkeys that are sympatric with mandrills. These observations lend support to the hypothesis that the primate lentiviruses originated and coevolved within monkeys of the Cercopithecus genus. Regarded in this light, lentivirus infections of primates not belonging to the Cercopithecus genus may have resulted from cross-species transmission in the not-too-distant past.
Figures
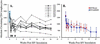

Similar articles
-
Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir.J Virol. 2000 Apr;74(8):3892-8. doi: 10.1128/jvi.74.8.3892-3898.2000. J Virol. 2000. PMID: 10729165 Free PMC article.
-
Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies.J Virol. 1999 Sep;73(9):7734-44. doi: 10.1128/JVI.73.9.7734-7744.1999. J Virol. 1999. PMID: 10438863 Free PMC article.
-
Wild Mandrillus sphinx are carriers of two types of lentivirus.J Virol. 2001 Aug;75(15):7086-96. doi: 10.1128/JVI.75.15.7086-7096.2001. J Virol. 2001. PMID: 11435589 Free PMC article.
-
Phylogeny and natural history of the primate lentiviruses, SIV and HIV.Curr Opin Genet Dev. 1995 Dec;5(6):798-806. doi: 10.1016/0959-437x(95)80014-v. Curr Opin Genet Dev. 1995. PMID: 8745080 Review.
-
The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa.Front Biosci. 2004 Jan 1;9:225-54. doi: 10.2741/1154. Front Biosci. 2004. PMID: 14766362 Review.
Cited by
-
Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti).J Virol. 2005 Jul;79(13):8560-71. doi: 10.1128/JVI.79.13.8560-8571.2005. J Virol. 2005. PMID: 15956597 Free PMC article.
-
Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir.J Virol. 2000 Apr;74(8):3892-8. doi: 10.1128/jvi.74.8.3892-3898.2000. J Virol. 2000. PMID: 10729165 Free PMC article.
-
Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus).J Virol. 2003 Jan;77(1):744-8. doi: 10.1128/jvi.77.1.744-748.2003. J Virol. 2003. PMID: 12477880 Free PMC article.
-
Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411).J Virol. 2001 Dec;75(24):12014-27. doi: 10.1128/JVI.75.24.12014-12027.2001. J Virol. 2001. PMID: 11711592 Free PMC article.
-
Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma.J Virol. 2003 Feb;77(3):2214-26. doi: 10.1128/jvi.77.3.2214-2226.2003. J Virol. 2003. PMID: 12525656 Free PMC article.
References
-
- Adachi J, Hasegawa M. MOLPHY (a program package for MOLecular PHYlogenetics), version 2.2. Tokyo, Japan: Institute of Statistical Mathematics; 1994.
-
- Allan J S. Pathogenic properties of simian immunodeficiency viruses in nonhuman primates. Annu Rev AIDS Res. 1991;1:191–206.
-
- Baier M, Werner A, Cichutek K, Garber C, Muller C, Kraus G, Ferdinand F J, Hartung S, Papas T S, Kurth R. Molecularly cloned simian immunodeficiency virus SIVagm3 is highly divergent from other SIVagm isolates and is biologically active in vitro and in vivo. J Virol. 1989;63:5119–5123. - PMC - PubMed
-
- Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado P, Galat G, Durant J P, Pourrut X, Veas F. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopthecus aethiops sabeus) in the wild. J Gen Virol. 1996;77:773–781. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

