The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3' untranslated region correlates with its affinity for the elav-like HuR protein
- PMID: 9882309
- PMCID: PMC103928
- DOI: 10.1128/JVI.73.2.1080-1091.1999
The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3' untranslated region correlates with its affinity for the elav-like HuR protein
Abstract
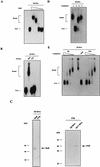
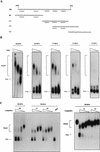
A 57-nucleotide adenosine- and uridine-rich RNA instability element in the human papillomavirus type 1 late 3' untranslated region termed h1ARE has previously been shown to interact specifically with three nuclear proteins that failed to bind to an inactive mutant RNA. Two of those were identified as the heterogeneous ribonucleoproteins C1 and C2, whereas the third, a 38-kDa, poly(U) binding protein (p38), remained unidentified. Here we show that partially purified p38 reacts with a monoclonal antibody raised against the recently identified elav-like HuR protein, indicating that p38 is the HuR protein. Indeed, recombinant glutathione S-transferase (GST)-HuR protein binds specifically to sites within the h1ARE. Determination of the apparent Kd value of GST-HuR for the h1ARE and the inactive mutant thereof revealed that GST-HuR bound with a more than 50-fold-higher affinity to the wild-type sequence. Therefore, the binding affinity of GST-HuR for the wild-type and mutant h1AREs correlates with their inhibitory activities in transfected cells, strongly suggesting that the HuR protein is involved in the posttranscriptional regulation of human papillomavirus type 1 late-gene expression.
Figures






Similar articles
-
Heterogeneous nuclear ribonucleoprotein C binds exclusively to the functionally important UUUUU-motifs in the human papillomavirus type-1 AU-rich inhibitory element.Virus Res. 2001 Mar;73(2):163-75. doi: 10.1016/s0168-1702(00)00238-0. Virus Res. 2001. PMID: 11172920
-
Inhibitory activity of the human papillomavirus type 1 AU-rich element correlates inversely with the levels of the elav-like HuR protein in the cell cytoplasm.Arch Virol. 2000;145(3):491-503. doi: 10.1007/s007050050041. Arch Virol. 2000. PMID: 10795517
-
HADHB, HuR, and CP1 bind to the distal 3'-untranslated region of human renin mRNA and differentially modulate renin expression.J Biol Chem. 2003 Nov 7;278(45):44894-903. doi: 10.1074/jbc.M307782200. Epub 2003 Aug 21. J Biol Chem. 2003. PMID: 12933794
-
HuR binding to AU-rich elements present in the 3' untranslated region of Classical swine fever virus.Virol J. 2011 Jul 6;8:340. doi: 10.1186/1743-422X-8-340. Virol J. 2011. PMID: 21729330 Free PMC article.
-
Properties of the Regulatory RNA-Binding Protein HuR and its Role in Controlling miRNA Repression.Adv Exp Med Biol. 2011;700:106-23. doi: 10.1007/978-1-4419-7823-3_10. Adv Exp Med Biol. 2011. PMID: 21755477 Review.
Cited by
-
Identifying regulatory elements and their RNA-binding proteins in the 3' untranslated regions of papillomavirus late mRNAs.Biomed Rep. 2024 Jul 1;21(2):125. doi: 10.3892/br.2024.1813. eCollection 2024 Aug. Biomed Rep. 2024. PMID: 39006509 Free PMC article.
-
HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells.Retrovirology. 2008 Jun 10;5:47. doi: 10.1186/1742-4690-5-47. Retrovirology. 2008. PMID: 18544151 Free PMC article.
-
Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies.Future Microbiol. 2010 Oct;5(10):1493-506. doi: 10.2217/fmb.10.107. Future Microbiol. 2010. PMID: 21073310 Free PMC article. Review.
-
Function of RRM domains of Drosophila melanogaster ELAV: Rnp1 mutations and rrm domain replacements with ELAV family proteins and SXL.Genetics. 2000 Aug;155(4):1789-98. doi: 10.1093/genetics/155.4.1789. Genetics. 2000. PMID: 10924474 Free PMC article.
-
The human papillomavirus type 31 late 3' untranslated region contains a complex bipartite negative regulatory element.J Virol. 2002 Jun;76(12):5993-6003. doi: 10.1128/jvi.76.12.5993-6003.2002. J Virol. 2002. PMID: 12021332 Free PMC article.
References
-
- Baker C, Calef C. Maps of papillomavirus mRNA transcripts. In: Billakanti S R, Calef C E, Farmer A D, Halpern A L, Myers G L, editors. Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Los Alamos National Laboratory; 1997. pp. 3–10.
-
- Baker C C. Post-transcriptional regulation of papillomavirus gene expression. In: Billakanti S R, Calef C E, Farmer A D, Halpern A L, Myers G L, editors. Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Los Alamos National Laboratory; 1997. pp. 11–16.
-
- Beelman C A, Parker R. Degradation of mRNA in eucaryotes. Cell. 1995;81:179–183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

