Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains
- PMID: 9882315
- PMCID: PMC103934
- DOI: 10.1128/JVI.73.2.1138-1145.1999
Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains
Abstract
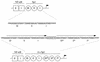
Live, attenuated viruses have been the most successful vaccines in monkey models of human immunodeficiency virus type 1 (HIV-1) infection. However, there are several safety concerns about using such an anti-HIV vaccine in humans, including reversion of the vaccine strain to virulence and recombination with endogenous retroviral sequences to produce new infectious and potentially pathogenic viruses. Because testing in humans would inevitably carry a substantial risk, we set out to test the genetic stability of multiply deleted HIV constructs in perpetuated tissue culture infections. The Delta3 candidate vaccine strain of HIV-1 contains deletions in the viral long terminal repeat (LTR) promoter and the vpr and nef genes. This virus replicates with delayed kinetics, but a profound enhancement of virus replication was observed after approximately 2 months of culturing. Analysis of the revertant viral genome indicated that the three introduced deletions were maintained but a 39-nucleotide sequence was inserted in the LTR promoter region. This insert was formed by duplication of the region encoding three binding sites for the Sp1 transcription factor. The duplicated Sp1 region was demonstrated to increase the LTR promoter activity, and a concomitant increase in the virus replication rate was measured. In fact, duplication of the Sp1 sites increased the fitness of the Delta3 virus (Vpr/Nef/U3) to levels higher than that of the singly deleted DeltaVpr virus. These results indicate that deleted HIV-1 vaccine strains can evolve into fast-replicating variants by multiplication of remaining sequence motifs, and their safety is therefore not guaranteed. This insight may guide future efforts to develop more stable anti-HIV vaccines.
Figures



References
-
- Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. - PubMed
-
- Baba T W, Jeong Y S, Penninck D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

