Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro
- PMID: 9882324
- PMCID: PMC103943
- DOI: 10.1128/JVI.73.2.1219-1226.1999
Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro
Abstract
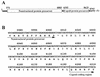
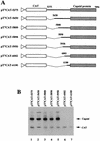
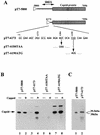
AUG-unrelated translation initiation was found in an insect picorna-like virus, Plautia stali intestine virus (PSIV). The positive-strand RNA genome of the virus contains two nonoverlapping open reading frames (ORFs). The capsid protein gene is located in the 3'-proximal ORF and lacks an AUG initiation codon. We examined the translation mechanism and the initiation codon of the capsid protein gene by using various dicistronic and monocistronic RNAs in vitro. The capsid protein gene was translated cap independently in the presence of the upstream cistron, indicating that the gene is translated by internal ribosome entry. Deletion analysis showed that the internal ribosome entry site (IRES) consisted of approximately 250 bases and that its 3' boundary extended slightly into the capsid-coding region. The initiation codon for the IRES-mediated translation was identified as the CUU codon, which is located just upstream of the 5' terminus of the capsid-coding region by site-directed mutagenesis. In vitro translation assays of monocistronic RNAs lacking the 5' part of the IRES showed that this CUU codon was not recognized by scanning ribosomes. This suggests that the PSIV IRES can effectively direct translation initiation without stable codon-anticodon pairing between the initiation codon and the initiator methionyl-tRNA.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

