Cloning and characterization of adeno-associated virus type 5
- PMID: 9882336
- PMCID: PMC103955
- DOI: 10.1128/JVI.73.2.1309-1319.1999
Cloning and characterization of adeno-associated virus type 5
Abstract
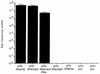
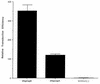
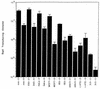
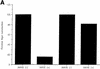
Adeno-associated virus type 5 (AAV5) is distinct from other dependovirus serotypes based on DNA hybridization and serological data. To better understand the biology of AAV5, we have cloned and sequenced its genome and generated recombinant AAV5 particles. The single-stranded DNA genome is similar in length and genetic organization to that of AAV2. The rep gene of AAV5 is 67% homologous to AAV2, with the majority of the changes occurring in the carboxyl and amino termini. This homology is much less than that observed with other reported AAV serotypes. The inverted terminal repeats (ITRs) are also unique compared to those of the other AAV serotypes. While the characteristic AAV hairpin structure and the Rep DNA binding site are retained, the consensus terminal resolution site is absent. These differences in the Rep proteins and the ITRs result in a lack of cross-complementation between AAV2 and AAV5 as measured by the production of recombinant AAV particles. Alignment of the cap open reading frame with that of the other AAV serotypes identifies both conserved and variable regions which could affect tissue tropism and particle stability. Comparison of transduction efficiencies in a variety of cells lines and a lack of inhibition by soluble heparin indicate that AAV5 may utilize a distinct mechanism of uptake compared to AAV2.
Figures














References
-
- Bantel-Schall U, zur Hausen H. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology. 1984;134:52–63. - PubMed
-
- Blacklow N R, Hoggan M D, Rowe W P. Serologic evidence for human infection with adeno-associated viruses. J Natl Cancer Inst. 1968;40:319–327. - PubMed
-
- Chapman M S, Rossmann M G. Structure, sequence, and function correlations among parvoviruses. Virology. 1993;194:491–508. - PubMed
-
- Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

