RNA-binding and capping activities of proteins in rotavirus open cores
- PMID: 9882343
- PMCID: PMC103962
- DOI: 10.1128/JVI.73.2.1382-1391.1999
RNA-binding and capping activities of proteins in rotavirus open cores
Abstract
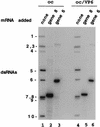
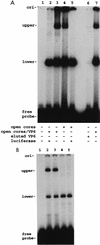
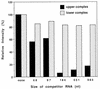
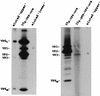
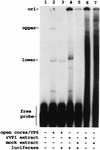
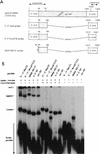
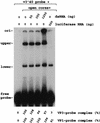
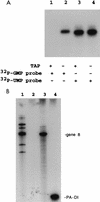
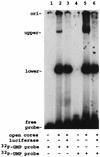
Guanylyltransferases are members of the nucleotidyltransferase family and function in mRNA capping by transferring GMP to the phosphate end of nascent RNAs. Although numerous guanylyltransferases have been identified, studies which define the nature of the interaction between the capping enzymes of any origin and their RNA substrates have been limited. Here, we have characterized the RNA-binding activity of VP3, a minor protein component of the core of rotavirions that has been proposed to function as the viral guanylyltransferase and to direct the capping of the 11 transcripts synthesized from the segmented double-stranded RNA (dsRNA) genome of these viruses. Gel shift analysis performed with disrupted (open) virion-derived cores and virus-specific RNA probes showed that VP3 has affinity for single-stranded RNA (ssRNA) but not for dsRNA. While the ssRNA-binding activity of VP3 was found to be sequence independent, the protein does exhibit preferential affinity for uncapped over capped RNA. Like the RNA-binding activity, RNA capping assays performed with open cores indicates that the guanylyltransferase activity of VP3 is nonspecific and is able to cap RNAs initiating with a G or an A residue. These data establish that all three rotavirus core proteins, VP1, the RNA polymerase; VP2, the core capsid protein; and VP3, the guanylyltransferase, have affinity for RNA but that only in the case of the RNA polymerase is the affinity sequence specific.
Figures



References
-
- Chen, D., and J. T. Patton. Unpublished data.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

