Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa
- PMID: 9882649
- PMCID: PMC93389
- DOI: 10.1128/JB.181.2.382-388.1999
Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa
Abstract
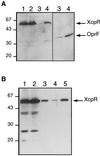
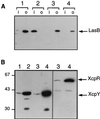
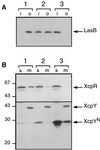
A broad range of extracellular proteins secreted by Pseudomonas aeruginosa use the type II or general secretory pathway (GSP) to reach the medium. This pathway requires the expression of at least 12 xcp gene products. XcpR, a putative nucleotide-binding protein, is essential for the secretion process across the outer membrane even though the protein contains no hydrophobic sequence that could target or anchor it to the bacterial envelope. For a better understanding of the relationship between XcpR and the other Xcp proteins which are located in the envelope, we have studied its subcellular localization. In a wild-type P. aeruginosa strain, XcpR was found associated with the cytoplasmic membrane. This association depends on the presence of the XcpY protein, which also appears to be necessary for XcpR stability. Functional complementation of an xcpY mutant required the XcpY protein to be expressed at a low level. Higher expression precluded the complementing activity of XcpY, although membrane association of XcpR was restored. This behavior suggested that an excess of free XcpY might interfere with the secretion by formation of inactive XcpR-XcpY complexes which cannot properly interact with their natural partners in the secretion machinery. These data show that a precise stoichiometric ratio between several components may be crucial for the functioning of the GSP.
Figures



References
-
- Akrim M, Bally M, Ball G, Tommassen J, Teerink H, Filloux A, Lazdunski A. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol Microbiol. 1993;10:431–443. - PubMed
-
- Bagdasarian M M, Amann E, Lurz R, Rückert B, Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983;26:273–282. - PubMed
-
- Ball, G., and M. Bally. Unpublished data.
-
- Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

