A glucan synthase FKS1 homolog in cryptococcus neoformans is single copy and encodes an essential function
- PMID: 9882657
- PMCID: PMC93397
- DOI: 10.1128/JB.181.2.444-453.1999
A glucan synthase FKS1 homolog in cryptococcus neoformans is single copy and encodes an essential function
Abstract
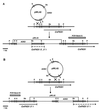
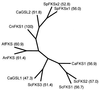
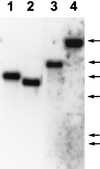
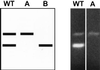
Cryptococcal meningitis is a fungal infection, caused by Cryptococcus neoformans, which is prevalent in immunocompromised patient populations. Treatment failures of this disease are emerging in the clinic, usually associated with long-term treatment with existing antifungal agents. The fungal cell wall is an attractive target for drug therapy because the syntheses of cell wall glucan and chitin are processes that are absent in mammalian cells. Echinocandins comprise a class of lipopeptide compounds known to inhibit 1,3-beta-glucan synthesis, and at least two compounds belonging to this class are currently in clinical trials as therapy for life-threatening fungal infections. Studies of Saccharomyces cerevisiae and Candida albicans mutants identify the membrane-spanning subunit of glucan synthase, encoded by the FKS genes, as the molecular target of echinocandins. In vitro, the echinocandins show potent antifungal activity against Candida and Aspergillus species but are much less potent against C. neoformans. In order to examine why C. neoformans cells are less susceptible to echinocandin treatment, we have cloned a homolog of S. cerevisiae FKS1 from C. neoformans. We have developed a generalized method to evaluate the essentiality of genes in Cryptococcus and applied it to the FKS1 gene. The method relies on homologous integrative transformation with a plasmid that can integrate in two orientations, only one of which will disrupt the target gene function. The results of this analysis suggest that the C. neoformans FKS1 gene is essential for viability. The C. neoformans FKS1 sequence is closely related to the FKS1 sequences from other fungal species and appears to be single copy in C. neoformans. Furthermore, amino acid residues known to be critical for echinocandin susceptibility in Saccharomyces are conserved in the C. neoformans FKS1 sequence.
Figures

Similar articles
-
Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase.Mol Cell Biol. 1995 Oct;15(10):5671-81. doi: 10.1128/MCB.15.10.5671. Mol Cell Biol. 1995. PMID: 7565718 Free PMC article.
-
Puf4 Mediates Post-transcriptional Regulation of Cell Wall Biosynthesis and Caspofungin Resistance in Cryptococcus neoformans.mBio. 2021 Jan 12;12(1):e03225-20. doi: 10.1128/mBio.03225-20. mBio. 2021. PMID: 33436441 Free PMC article.
-
Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis.J Bacteriol. 1997 Jul;179(13):4096-105. doi: 10.1128/jb.179.13.4096-4105.1997. J Bacteriol. 1997. PMID: 9209021 Free PMC article.
-
Lipopeptide inhibitors of fungal glucan synthase.J Med Vet Mycol. 1997 Mar-Apr;35(2):79-86. doi: 10.1080/02681219780000961. J Med Vet Mycol. 1997. PMID: 9147267 Review.
-
Resistance to echinocandin-class antifungal drugs.Drug Resist Updat. 2007 Jun;10(3):121-30. doi: 10.1016/j.drup.2007.04.002. Epub 2007 Jun 13. Drug Resist Updat. 2007. PMID: 17569573 Free PMC article. Review.
Cited by
-
Will the Real Immunogens Please Stand Up: Exploiting the Immunogenic Potential of Cryptococcal Cell Antigens in Fungal Vaccine Development.J Fungi (Basel). 2024 Dec 4;10(12):840. doi: 10.3390/jof10120840. J Fungi (Basel). 2024. PMID: 39728336 Free PMC article. Review.
-
Adaptation of the tetracycline-repressible system for modulating the expression of essential genes in Cryptococcus neoformans.mSphere. 2025 May 27;10(5):e0101824. doi: 10.1128/msphere.01018-24. Epub 2025 May 1. mSphere. 2025. PMID: 40310102 Free PMC article.
-
Cell wall assembly in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2006 Jun;70(2):317-43. doi: 10.1128/MMBR.00038-05. Microbiol Mol Biol Rev. 2006. PMID: 16760306 Free PMC article. Review.
-
Combating increased antifungal drug resistance in Cryptococcus, what should we do in the future?Acta Biochim Biophys Sin (Shanghai). 2023 Feb 22;55(4):540-547. doi: 10.3724/abbs.2023011. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 36815374 Free PMC article. Review.
-
Ergosterol distribution controls surface structure formation and fungal pathogenicity.bioRxiv [Preprint]. 2023 Feb 17:2023.02.17.528979. doi: 10.1101/2023.02.17.528979. bioRxiv. 2023. Update in: mBio. 2023 Aug 31;14(4):e0135323. doi: 10.1128/mbio.01353-23. PMID: 36824733 Free PMC article. Updated. Preprint.
References
-
- Bartizal K, Abruzzo G, Trainor C, Krupa D, Nollstadt K, Schmatz D, Schwartz R, Hammond M, Balkovec J, Vanmiddlesworth F. In vitro antifungal activities and in vivo efficacies of 1,3-β-d-glucan synthesis inhibitors L-671,329, L-646,991, tetrahydroechinocandin B, and L-687,781, a papulacandin. Antimicrob Agents Chemother. 1992;36:1648–1657. - PMC - PubMed
-
- Bastide J M, Hadibi E H, Bastide M. Taxonomic significance of yeast sphaeroplast release after enzymatic treatment of intact cells. J Gen Microbiol. 1979;113:147–153. - PubMed
-
- Berg J, Clancy C J, Nguyen M H. The hidden danger of primary fluconazole prophylaxis for patients with AIDS. Clin Infect Dis. 1998;26:186–187. - PubMed
-
- Bouffard F A, Zambias R A, Dropinski J F, Balkovec J M, Hammond M L, Abruzzo G K, Bartizal K F, Marrinan J A, Kurtz M B, McFadden D C, Nollstadt K H, Powles M A, Schmatz D M. Synthesis and antifungal activity of novel cationic pneumocandin B0 derivatives. J Med Chem. 1994;37:222–225. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

