Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast
- PMID: 9885246
- PMCID: PMC2148128
- DOI: 10.1083/jcb.144.1.83
Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast
Abstract
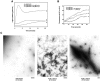
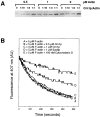
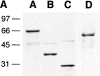
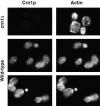
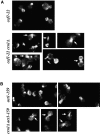
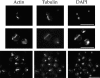
Coronin is a highly conserved actin-associated protein that until now has had unknown biochemical activities. Using microtubule affinity chromatography, we coisolated actin and a homologue of coronin, Crn1p, from Saccharomyces cerevisiae cell extracts. Crn1p is an abundant component of the cortical actin cytoskeleton and binds to F-actin with high affinity (Kd 6 x 10(-9) M). Crn1p promotes the rapid barbed-end assembly of actin filaments and cross-links filaments into bundles and more complex networks, but does not stabilize them. Genetic analyses with a crn1Delta deletion mutation also are consistent with Crn1p regulating filament assembly rather than stability. Filament cross-linking depends on the coiled coil domain of Crn1p, suggesting a requirement for Crn1p dimerization. Assembly-promoting activity is independent of cross-linking and could be due to nucleation and/or accelerated polymerization. Crn1p also binds to microtubules in vitro, and microtubule binding is enhanced by the presence of actin filaments. Microtubule binding is mediated by a region of Crn1p that contains sequences (not found in other coronins) homologous to the microtubule binding region of MAP1B. These activities, considered with microtubule defects observed in crn1Delta cells and in cells overexpressing Crn1p, suggest that Crn1p may provide a functional link between the actin and microtubule cytoskeletons in yeast.
Figures






References
-
- Ayscough, K.R., and D.G. Drubin. 1997. Immunofluorescence microscopy of yeast cells. In Cell Biology, A Laboratory Handbook. Academic Press. San Diego, CA.
-
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

