A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation
- PMID: 9891073
- PMCID: PMC116068
- DOI: 10.1128/MCB.19.2.1401
A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation
Abstract
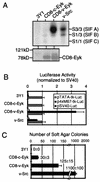
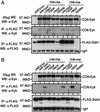
The receptor tyrosine kinase Eyk, a member of the Axl/Tyro3 subfamily, activates the STAT pathway and transforms cells when constitutively activated. Here, we compared the potentials of the intracellular domains of Eyk molecules derived from c-Eyk and v-Eyk to transform rat 3Y1 fibroblasts. The v-Eyk molecule induced higher numbers of transformants in soft agar and stronger activation of Stat3; levels of Stat1 activation by the two Eyk molecules were similar. A mutation in the sequence Y933VPL, present in c-Eyk, to the v-Eyk sequence Y933VPQ led to increased activation of Stat3 and increased transformation efficiency. However, altering another sequence, Y862VNT, present in both Eyk molecules to F862VNT markedly decreased transformation without impairing Stat3 activation. These results indicate that activation of Stat3 enhances transformation efficiency and cooperates with another pathway to induce transformation.
Figures



References
-
- Bellosta P, Zhang Q, Goff S P, Basilico C. Signaling through the ARK tyrosine kinase receptor protects from apoptosis in the absence of growth stimulation. Oncogene. 1997;15:2387–2397. - PubMed
-
- Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. - PubMed
-
- Biscardi J S, Denhez F, Buehler G F, Chesnutt D A, Baragona S C, O’Bryan J P, Der C J, Fiordalisi J J, Fults D W, Maness P F. Rek, a gene expressed in retina and brain, encodes a receptor tyrosine kinase of the Axl/Tyro3 family. J Biol Chem. 1996;271:29049–29059. - PubMed
-
- Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
