Persistence of an alternate chromatin structure at silenced loci in vitro
- PMID: 9892635
- PMCID: PMC15138
- DOI: 10.1073/pnas.96.2.343
Persistence of an alternate chromatin structure at silenced loci in vitro
Abstract
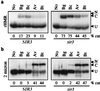
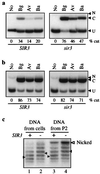
In Saccharomyces cerevisiae, transcriptional repression at the HM mating-type loci and telomeres results from the formation of a heterochromatin-like structure. Silencing requires at least three Sir proteins (Sir2p-4p), which are recruited to chromatin by silencers at the HM loci and TG1-3 tracts at telomeres. Sir proteins and telomeres colocalize at the nuclear periphery, suggesting that this subnuclear position may also contribute to transcriptional repression. To evaluate the contribution of nuclear context to silencing, we developed methodology to isolate silent chromatin for analysis in vitro. Site-specific recombination was used in vivo to produce DNA rings from the silent HMR locus, and differential centrifugation was used to isolate the rings from whole-cell lysate. The partially purified rings retained many of the intracellular hallmarks of transcriptionally repressed domains. Specifically, rings from repressed strains were resistant to restriction endonuclease digestion, bore an altered DNA topology, and were associated with Sir3p. The recombination approach also was used to form rings from HMR that lacked silencers. Despite the uncoupling of these cis-acting regulatory elements, similar but nonidentical results were obtained. We conclude that an alternate chromatin structure at silent loci can persist in vitro in the absence of silencers and nuclear compartmentalization.
Figures




Similar articles
-
Persistence of an alternate chromatin structure at silenced loci in the absence of silencers.Proc Natl Acad Sci U S A. 1998 May 12;95(10):5521-6. doi: 10.1073/pnas.95.10.5521. Proc Natl Acad Sci U S A. 1998. PMID: 9576915 Free PMC article.
-
Perinuclear localization of chromatin facilitates transcriptional silencing.Nature. 1998 Aug 6;394(6693):592-5. doi: 10.1038/29100. Nature. 1998. PMID: 9707122
-
Yeast heterochromatin is a dynamic structure that requires silencers continuously.Genes Dev. 2000 Feb 15;14(4):452-63. Genes Dev. 2000. PMID: 10691737 Free PMC article.
-
Silent chromatin in yeast: an orchestrated medley featuring Sir3p [corrected].Bioessays. 1998 Jan;20(1):30-40. doi: 10.1002/(SICI)1521-1878(199801)20:1<30::AID-BIES6>3.0.CO;2-W. Bioessays. 1998. PMID: 9504045 Review.
-
Silencers, silencing, and heritable transcriptional states.Microbiol Rev. 1992 Dec;56(4):543-60. doi: 10.1128/mr.56.4.543-560.1992. Microbiol Rev. 1992. PMID: 1480108 Free PMC article. Review.
Cited by
-
Impact of Homologous Recombination on Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2018 Mar;208(3):1099-1113. doi: 10.1534/genetics.118.300704. Epub 2018 Jan 16. Genetics. 2018. PMID: 29339409 Free PMC article.
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
-
A single heterochromatin boundary element imposes position-independent antisilencing activity in Saccharomyces cerevisiae minichromosomes.PLoS One. 2011;6(9):e24835. doi: 10.1371/journal.pone.0024835. Epub 2011 Sep 16. PLoS One. 2011. PMID: 21949764 Free PMC article.
-
Yeast heterochromatin regulators Sir2 and Sir3 act directly at euchromatic DNA replication origins.PLoS Genet. 2018 May 24;14(5):e1007418. doi: 10.1371/journal.pgen.1007418. eCollection 2018 May. PLoS Genet. 2018. PMID: 29795547 Free PMC article.
-
Long-range communication between the silencers of HMR.Mol Cell Biol. 2008 Mar;28(6):1924-35. doi: 10.1128/MCB.01647-07. Epub 2008 Jan 14. Mol Cell Biol. 2008. PMID: 18195043 Free PMC article.
References
-
- Elgin S C R. Curr Opin Genet Dev. 1996;6:193–202. - PubMed
-
- Csink A K, Henikoff S. Nature (London) 1996;381:529–531. - PubMed
-
- Dernburg A F, Broman K W, Fung J C, Marshall W F, Philips J, Agard D A, Sedat J W. Cell. 1996;85:745–759. - PubMed
-
- Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. - PubMed
-
- Hecht A, Strahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

