Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies
- PMID: 9892643
- PMCID: PMC15146
- DOI: 10.1073/pnas.96.2.388
Chemical ligation of folded recombinant proteins: segmental isotopic labeling of domains for NMR studies
Abstract
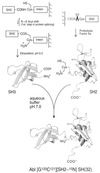
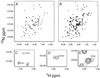
A convenient in vitro chemical ligation strategy has been developed that allows folded recombinant proteins to be joined together. This strategy permits segmental, selective isotopic labeling of the product. The src homology type 3 and 2 domains (SH3 and SH2) of Abelson protein tyrosine kinase, which constitute the regulatory apparatus of the protein, were individually prepared in reactive forms that can be ligated together under normal protein-folding conditions to form a normal peptide bond at the ligation junction. This strategy was used to prepare NMR sample quantities of the Abelson protein tyrosine kinase-SH(32) domain pair, in which only one of the domains was labeled with 15N. Mass spectrometry and NMR analyses were used to confirm the structure of the ligated protein, which was also shown to have appropriate ligand-binding properties. The ability to prepare recombinant proteins with selectively labeled segments having a single-site mutation, by using a combination of expression of fusion proteins and chemical ligation in vitro, will increase the size limits for protein structural determination in solution with NMR methods. In vitro chemical ligation of expressed protein domains will also provide a combinatorial approach to the synthesis of linked protein domains.
Figures

Comment in
-
Extending the size limit of protein nuclear magnetic resonance.Proc Natl Acad Sci U S A. 1999 Jan 19;96(2):332-4. doi: 10.1073/pnas.96.2.332. Proc Natl Acad Sci U S A. 1999. PMID: 9892632 Free PMC article. No abstract available.
Similar articles
-
The solution structure of Abl SH3, and its relationship to SH2 in the SH(32) construct.Structure. 1995 Oct 15;3(10):1075-86. doi: 10.1016/s0969-2126(01)00243-x. Structure. 1995. PMID: 8590002
-
Segmental isotopic labeling of proteins for nuclear magnetic resonance.Methods Enzymol. 2009;462:151-75. doi: 10.1016/S0076-6879(09)62008-5. Methods Enzymol. 2009. PMID: 19632474 Free PMC article.
-
1H and 15N assignments and secondary structure of the Src SH3 domain.FEBS Lett. 1993 Jun 7;324(1):87-92. doi: 10.1016/0014-5793(93)81538-b. FEBS Lett. 1993. PMID: 8504863
-
Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14053-8. doi: 10.1073/pnas.212518799. Epub 2002 Oct 16. Proc Natl Acad Sci U S A. 2002. PMID: 12384576 Free PMC article.
-
Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.J Mol Biol. 1998 Feb 20;276(2):461-78. doi: 10.1006/jmbi.1997.1562. J Mol Biol. 1998. PMID: 9512716
Cited by
-
The dynamic duo: combining NMR and small angle scattering in structural biology.Protein Sci. 2014 Jun;23(6):669-82. doi: 10.1002/pro.2467. Epub 2014 Apr 17. Protein Sci. 2014. PMID: 24687405 Free PMC article. Review.
-
Expressed protein ligation using an N-terminal cysteine containing fragment generated in vivo from a pelB fusion protein.Protein Expr Purif. 2007 Aug;54(2):227-33. doi: 10.1016/j.pep.2007.04.002. Epub 2007 Apr 10. Protein Expr Purif. 2007. PMID: 17493830 Free PMC article.
-
Extending the size limit of protein nuclear magnetic resonance.Proc Natl Acad Sci U S A. 1999 Jan 19;96(2):332-4. doi: 10.1073/pnas.96.2.332. Proc Natl Acad Sci U S A. 1999. PMID: 9892632 Free PMC article. No abstract available.
-
Simultaneous detection and deconvolution of congested NMR spectra containing three isotopically labeled species.J Am Chem Soc. 2008 Jun 25;130(25):7818-9. doi: 10.1021/ja802701w. Epub 2008 May 31. J Am Chem Soc. 2008. PMID: 18512910 Free PMC article.
-
Split Inteins: Nature's Protein Ligases.Isr J Chem. 2011 Nov 1;51(8-9):854-861. doi: 10.1002/ijch.201100094. Isr J Chem. 2011. PMID: 23620603 Free PMC article.
References
-
- Jaenicke R. Biochemistry. 1991;30:3147–3161. - PubMed
-
- Bork P, Schultz J, Ponting C P. Trends Biochem Sci. 1997;22:296–298. - PubMed
-
- Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986.
-
- Clore, G. M. & Gronenborn, A. M. (1997) Nat. Struct. Biol. 4, Suppl., 849–853. - PubMed
-
- Wuthrich, K. (1998) Nat. Struct. Biol. 5, Suppl., 492–495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

