Morphological, physiological, and biochemical changes in rhodopsin knockout mice
- PMID: 9892703
- PMCID: PMC15206
- DOI: 10.1073/pnas.96.2.736
Morphological, physiological, and biochemical changes in rhodopsin knockout mice
Abstract
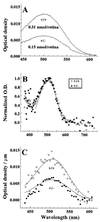
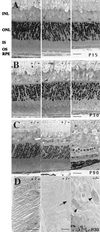
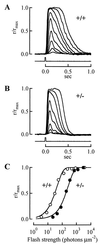
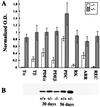
Mutations in rod opsin, the visual pigment protein of rod photoreceptors, account for approximately 15% of all inherited human retinal degenerations. However, the physiological and molecular events underlying the disease process are not well understood. One approach to this question has been to study transgenic mice expressing opsin genes containing defined mutations. A caveat of this approach is that even the overexpression of normal opsin leads to photoreceptor cell degeneration. To overcome the problem, we have reduced or eliminated endogenous rod opsin content by targeted gene disruption. Retinas in mice lacking both opsin alleles initially developed normally, except that rod outer segments failed to form. Within months of birth, photoreceptor cells degenerated completely. Retinas from mice with a single copy of the opsin gene developed normally, and rods elaborated outer segments of normal size but with half the normal complement of rhodopsin. Photoreceptor cells in these retinas also degenerated but did so over a much slower time course. Physiological and biochemical experiments showed that rods from mice with a single opsin gene were approximately 50% less sensitive to light, had accelerated flash-response kinetics, and contained approximately 50% more phosducin than wild-type controls.
Figures
References
-
- Gal A, Apfelstedt-Sylla E, Janecke A R, Zrenner E. Prog Retin Eye Res. 1997;16:51–79.
-
- Chen J, Makino C L, Peachey N S, Baylor D A, Simon M I. Science. 1995;267:374–377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

