Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin
- PMID: 9916102
- PMCID: PMC96398
- DOI: 10.1128/IAI.67.2.871-878.1999
Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin
Abstract
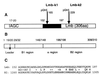
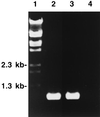
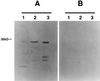
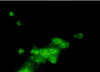
Streptococcus agalactiae is a leading cause of neonatal sepsis and meningitis. Adherence to extracellular matrix proteins is considered an important factor in the pathogenesis of infection, but the genetic determinants of this process remain largely unknown. We identified and sequenced a gene which codes for a putative lipoprotein that exhibits significant homology to the streptococcal LraI protein family. Mutants of this locus were demonstrated to have substantially reduced adherence to immobilized human laminin. The nucleotide sequence of the gene was subsequently designated lmb (laminin binding) and shown to be present in all of the common serotypes of S. agalactiae. To determine the role of Lmb in the adhesion of S. agalactiae wild-type strains to laminin, a recombinant Lmb protein harboring six consecutive histidine residues at the C terminus was cloned, expressed, and purified from Escherichia coli. Preincubation of immobilized laminin with recombinant Lmb significantly reduced adherence of the wild-type strain O90R to laminin. These results indicate that Lmb mediates the attachment of S. agalactiae to human laminin, which may be essential for the bacterial colonization of damaged epithelium and translocation of bacteria into the bloodstream.
Figures


Similar articles
-
Enhanced expression of lmb gene encoding laminin-binding protein in Streptococcus agalactiae strains harboring IS1548 in scpB-lmb intergenic region.PLoS One. 2010 May 24;5(5):e10794. doi: 10.1371/journal.pone.0010794. PLoS One. 2010. PMID: 20520730 Free PMC article.
-
Streptococcus agalactiae invasion of human brain microvascular endothelial cells is promoted by the laminin-binding protein Lmb.Microbes Infect. 2007 May;9(6):714-20. doi: 10.1016/j.micinf.2007.02.015. Epub 2007 Feb 24. Microbes Infect. 2007. PMID: 17400016
-
Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb.Mol Microbiol. 2001 Aug;41(4):925-35. doi: 10.1046/j.1365-2958.2001.02563.x. Mol Microbiol. 2001. PMID: 11532154
-
Collagen-like proteins of pathogenic streptococci.Mol Microbiol. 2017 Mar;103(6):919-930. doi: 10.1111/mmi.13604. Epub 2017 Jan 18. Mol Microbiol. 2017. PMID: 27997716 Free PMC article. Review.
-
Physiological impact of transposable elements encoding DDE transposases in the environmental adaptation of Streptococcus agalactiae.Microbiology (Reading). 2014 Jul;160(Pt 7):1298-1315. doi: 10.1099/mic.0.077628-0. Epub 2014 Apr 23. Microbiology (Reading). 2014. PMID: 24760965 Review.
Cited by
-
A Novel Conserved Protein in Streptococcus agalactiae, BvaP, Is Important for Vaginal Colonization and Biofilm Formation.mSphere. 2022 Dec 21;7(6):e0042122. doi: 10.1128/msphere.00421-22. Epub 2022 Oct 11. mSphere. 2022. PMID: 36218343 Free PMC article.
-
Genotypes and virulence genes in group B streptococcus isolated in the maternity hospital, Kuwait.Med Princ Pract. 2013;22(5):453-7. doi: 10.1159/000349932. Epub 2013 Apr 5. Med Princ Pract. 2013. PMID: 23571853 Free PMC article.
-
Human MOSPD2: A bacterial Lmb mimicked auto-antigen is involved in immune infertility.J Transl Autoimmun. 2019 May 28;1:100002. doi: 10.1016/j.jtauto.2019.100002. eCollection 2019 Apr. J Transl Autoimmun. 2019. PMID: 32743492 Free PMC article.
-
Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes.Infect Immun. 2007 Mar;75(3):1310-7. doi: 10.1128/IAI.00996-06. Epub 2006 Dec 11. Infect Immun. 2007. PMID: 17158903 Free PMC article.
-
Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence.Infect Immun. 2000 Aug;68(8):4441-51. doi: 10.1128/IAI.68.8.4441-4451.2000. Infect Immun. 2000. PMID: 10899841 Free PMC article.
References
-
- Bayles K W. The use of degenerate, sensor gene specific, oligodeoxyribonucleotide primers to amplify DNA fragments from Staphylococcus aureus. Gene. 1993;123:99–103. - PubMed
-
- Dintilhac A, Alloing G, Granadel C, Claverys J P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

