Multiple mutation analyses in single tumor cells with improved whole genome amplification
- PMID: 9916922
- PMCID: PMC1853424
- DOI: 10.1016/S0002-9440(10)65254-6
Multiple mutation analyses in single tumor cells with improved whole genome amplification
Abstract
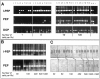
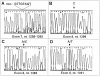
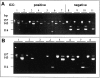
Combining whole genome amplification (WGA) methods with novel laser-based microdissection techniques has made it possible to exploit recent progress in molecular knowledge of cancer development and progression. However, WGA of one or a few cells has not yet been optimized and systematically evaluated for samples routinely processed in tumor pathology. We therefore studied the value of established WGA protocols in comparison to an improved PEP (I-PEP) PCR method in defined numbers of flow-sorted and microdissected tumor cells obtained both from frozen as well as formalin-fixed and paraffin-embedded tissue sections. In addition, the feasibility of I-PEP-PCR for mutation analysis was tested using clusters of 50-100 unfixed tumor cells obtained by touch preparation of ten breast carcinomas by conventional sequencing of exon 7 and 8 of the p53 gene. Finally, immunocytochemically stained microdissected single disseminated tumor cells from bone marrow aspirates were investigated with respect to mutations in codon 12 of Ki-ras by restriction fragment length polymorphism (RFLP)-PCR after I-PEP-PCR. The modified I-PEP-PCR protocol was superior to the original PEP-PCR and DOP-PCR protocols concerning amplification of DNA from one cell (efficiency rate I-PEP-PCR 40% versus PEP-PCR 15% and DOP-PCR 30%) and five cells (efficiency rate I-PEP-PCR 100% versus PEP-PCR 33% and DOP-PCR 20%). Preamplification by I-PEP allowed 100% sequence accuracy in > 4000 sequenced base pairs and Ki-ras mutation detection in isolated single disseminated tumor cells. For reliable microsatellite analysis of I-PEP-preamplified DNA, at least 10 unfixed cells from fluorescence-activated cell sorting, 10 cells from frozen tissue, or at least 30 cells from formalin-fixed and paraffin-embedded tissue sections were required. Thus, I-PEP-PCR allowed multiple reliable microsatellite analyses suited for microsatellite instability and losses of heterozygosity and mutation analysis even at the single cell level, rendering this technique a powerful new tool for molecular analyses in diagnostic and experimental tumor pathology.
Figures

References
-
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA: Laser capture microdissection. Science 1996, 274:998-1001 - PubMed
-
- Boehm M, Wieland I: Analysis of tumor-specific alterations in native specimens by PCR: how to procure the tumour cells. Int J Oncology 1997, 10:131-139 - PubMed
-
- Barret MT, Galipeau PC, Sanchez CA, Emond MJ, Reid BJ: Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene 1996, 12:1873-1878 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

