Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi
- PMID: 9922254
- PMCID: PMC93457
- DOI: 10.1128/JB.181.3.899-906.1999
Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi
Abstract
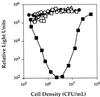
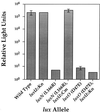
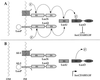
Vibrio harveyi regulates the expression of bioluminescence (lux) in response to cell density, a phenomenon known as quorum sensing. In V. harveyi, two independent quorum-sensing systems exist, and each produces, detects, and responds to a specific cell density-dependent autoinducer signal. The autoinducers are recognized by two-component hybrid sensor kinases called LuxN and LuxQ, and sensory information from both systems is transduced by a phosphorelay mechanism to the response regulator protein LuxO. Genetic evidence suggests that LuxO-phosphate negatively regulates the expression of luminescence at low cell density in the absence of autoinducers. At high cell density, interaction of the sensors with their cognate autoinducers results in dephosphorylation and inactivation of the LuxO repressor. In the present report, we show that LuxN and LuxQ channel sensory information to LuxO via a newly identified phosphorelay protein that we have named LuxU. LuxU shows sequence similarity to other described phosphorelay proteins, including BvgS, ArcB, and Ypd1. A critical His residue (His 58) of LuxU is required for phosphorelay function.
Figures


References
-
- Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. - PubMed
-
- Bainton N J, Bycroft B W, Chhabra S, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116:87–91. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

